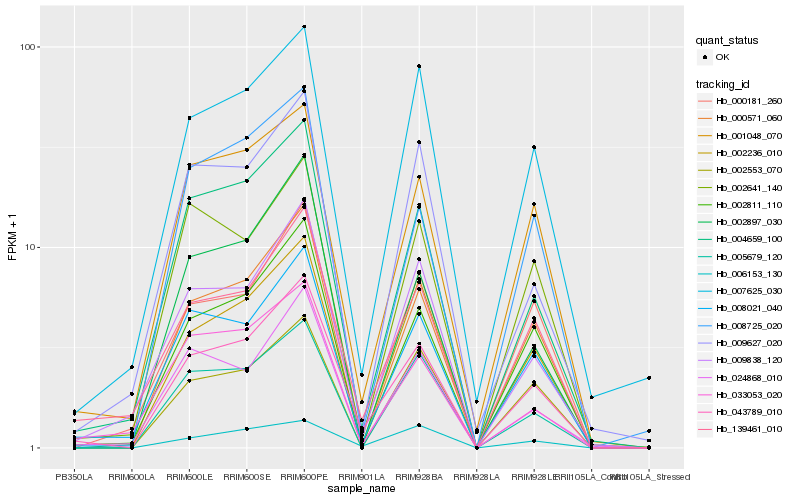
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_043789_010 |
0.0 |
transcription factor |
TF Family: AP2 |
hypothetical protein VITISV_034131 [Vitis vinifera] |
| 2 |
Hb_139461_010 |
0.0980334531 |
desease resistance |
Gene Name: NB-ARC |
Cc-nbs-lrr resistance protein, putative [Theobroma cacao] |
| 3 |
Hb_002811_110 |
0.1053991988 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 4 |
Hb_008021_040 |
0.1057445489 |
- |
- |
PREDICTED: AP2-like ethylene-responsive transcription factor ANT [Jatropha curcas] |
| 5 |
Hb_004659_100 |
0.1066882694 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000571_060 |
0.1085931157 |
- |
- |
PREDICTED: uncharacterized protein LOC105648112 [Jatropha curcas] |
| 7 |
Hb_001048_070 |
0.1151994622 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0007s14590g [Populus trichocarpa] |
| 8 |
Hb_002236_010 |
0.121419467 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET4 [Jatropha curcas] |
| 9 |
Hb_000181_260 |
0.1236368003 |
- |
- |
PREDICTED: uncharacterized protein LOC105139156 [Populus euphratica] |
| 10 |
Hb_009838_120 |
0.1251564292 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA13 isoform X1 [Jatropha curcas] |
| 11 |
Hb_024868_010 |
0.1272565956 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 12 |
Hb_007625_030 |
0.131396723 |
- |
- |
unknown [Medicago truncatula] |
| 13 |
Hb_005679_120 |
0.13355833 |
- |
- |
hypothetical protein RCOM_0284080 [Ricinus communis] |
| 14 |
Hb_002897_030 |
0.1346762975 |
- |
- |
PREDICTED: methylesterase 17 [Jatropha curcas] |
| 15 |
Hb_008725_020 |
0.1356470352 |
- |
- |
PREDICTED: cytidine deaminase 1 [Jatropha curcas] |
| 16 |
Hb_006153_130 |
0.1393211678 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 17 |
Hb_002641_140 |
0.1416013117 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 18 |
Hb_033053_020 |
0.1423252743 |
- |
- |
hypothetical protein JCGZ_01204 [Jatropha curcas] |
| 19 |
Hb_002553_070 |
0.1437008385 |
- |
- |
PREDICTED: RING-H2 finger protein ATL16 [Jatropha curcas] |
| 20 |
Hb_009627_020 |
0.1441965266 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 34 [Jatropha curcas] |