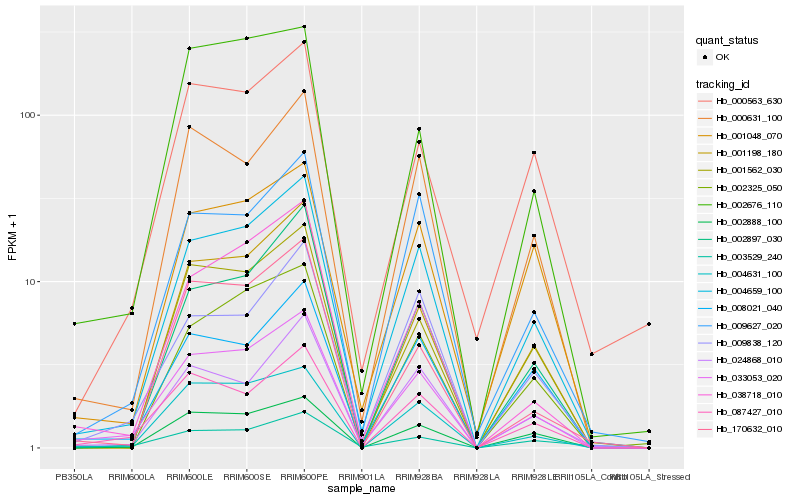
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033053_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_01204 [Jatropha curcas] |
| 2 |
Hb_004659_100 |
0.0718166995 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_087427_010 |
0.0950174309 |
- |
- |
hypothetical protein PRUPE_ppa007748mg [Prunus persica] |
| 4 |
Hb_004631_100 |
0.0997433708 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH96 [Jatropha curcas] |
| 5 |
Hb_000631_100 |
0.1137818109 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3-like [Jatropha curcas] |
| 6 |
Hb_038718_010 |
0.1175609614 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 7 |
Hb_008021_040 |
0.1188023282 |
- |
- |
PREDICTED: AP2-like ethylene-responsive transcription factor ANT [Jatropha curcas] |
| 8 |
Hb_002888_100 |
0.1197062791 |
- |
- |
PREDICTED: uncharacterized protein LOC103699074, partial [Phoenix dactylifera] |
| 9 |
Hb_009627_020 |
0.1230513407 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 34 [Jatropha curcas] |
| 10 |
Hb_024868_010 |
0.1236647781 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 11 |
Hb_009838_120 |
0.1245082049 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA13 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001198_180 |
0.125813764 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 13 |
Hb_001562_030 |
0.1261667107 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH35 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003529_240 |
0.1277466948 |
transcription factor |
TF Family: AP2 |
PREDICTED: ethylene-responsive transcription factor RAP2-7-like isoform X2 [Gossypium raimondii] |
| 15 |
Hb_002897_030 |
0.1295352553 |
- |
- |
PREDICTED: methylesterase 17 [Jatropha curcas] |
| 16 |
Hb_002325_050 |
0.1301035653 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus nigra] |
| 17 |
Hb_002676_110 |
0.1315614091 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase 2-like [Gossypium raimondii] |
| 18 |
Hb_001048_070 |
0.1349456428 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0007s14590g [Populus trichocarpa] |
| 19 |
Hb_170632_010 |
0.1372322514 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 20 |
Hb_000563_630 |
0.138724889 |
- |
- |
PREDICTED: uncharacterized protein LOC105643442 [Jatropha curcas] |