| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
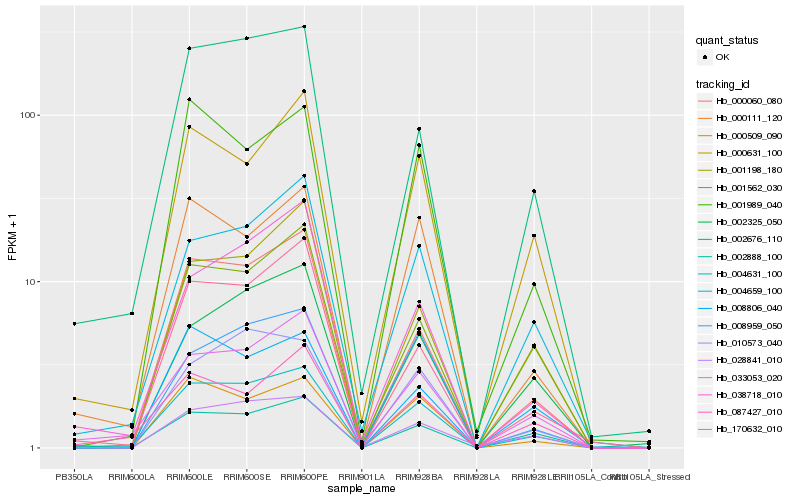
Hb_004631_100 |
0.0 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH96 [Jatropha curcas] |
| 2 |
Hb_002676_110 |
0.0979791592 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase 2-like [Gossypium raimondii] |
| 3 |
Hb_033053_020 |
0.0997433708 |
- |
- |
hypothetical protein JCGZ_01204 [Jatropha curcas] |
| 4 |
Hb_000060_080 |
0.1052627245 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 2 protein, putative [Ricinus communis] |
| 5 |
Hb_002888_100 |
0.1087456942 |
- |
- |
PREDICTED: uncharacterized protein LOC103699074, partial [Phoenix dactylifera] |
| 6 |
Hb_004659_100 |
0.1108981889 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000111_120 |
0.1110891685 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 8 |
Hb_000631_100 |
0.1168383451 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3-like [Jatropha curcas] |
| 9 |
Hb_028841_010 |
0.117436058 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase CES101 [Jatropha curcas] |
| 10 |
Hb_000509_090 |
0.1178895268 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 11 |
Hb_170632_010 |
0.1225046297 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 12 |
Hb_001562_030 |
0.1241626549 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH35 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001989_040 |
0.1268576785 |
- |
- |
PREDICTED: neurofilament heavy polypeptide [Jatropha curcas] |
| 14 |
Hb_002325_050 |
0.1307645058 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus nigra] |
| 15 |
Hb_010573_040 |
0.1311518469 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal isoform X2 [Populus euphratica] |
| 16 |
Hb_087427_010 |
0.1332666054 |
- |
- |
hypothetical protein PRUPE_ppa007748mg [Prunus persica] |
| 17 |
Hb_001198_180 |
0.1372710892 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 18 |
Hb_008959_050 |
0.1390896939 |
transcription factor |
TF Family: Orphans |
PREDICTED: late secretory pathway protein AVL9-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_008806_040 |
0.1416270792 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 20 |
Hb_038718_010 |
0.1416773765 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |