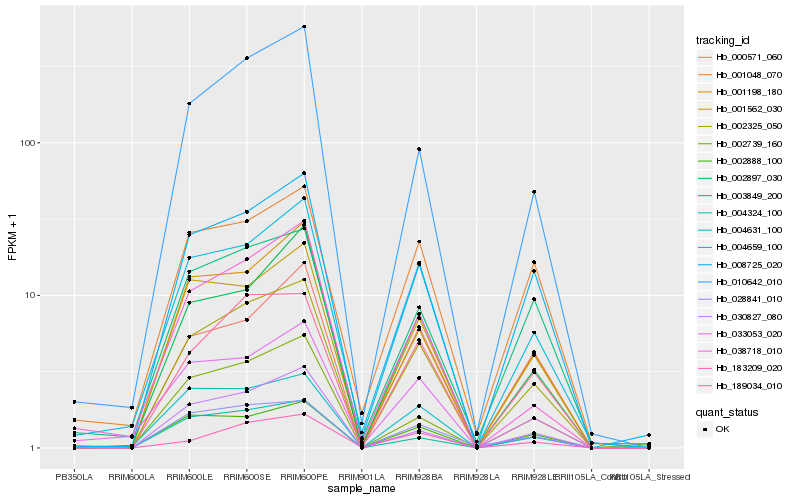
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002325_050 |
0.0 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus nigra] |
| 2 |
Hb_008725_020 |
0.0829722883 |
- |
- |
PREDICTED: cytidine deaminase 1 [Jatropha curcas] |
| 3 |
Hb_189034_010 |
0.0933440617 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_004659_100 |
0.0978916673 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001198_180 |
0.1019437767 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 6 |
Hb_010642_010 |
0.1052756992 |
- |
- |
PREDICTED: aquaporin TIP2-1 [Jatropha curcas] |
| 7 |
Hb_001562_030 |
0.1165481437 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH35 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002888_100 |
0.1177208543 |
- |
- |
PREDICTED: uncharacterized protein LOC103699074, partial [Phoenix dactylifera] |
| 9 |
Hb_003849_200 |
0.1206490824 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 10 |
Hb_002897_030 |
0.120830079 |
- |
- |
PREDICTED: methylesterase 17 [Jatropha curcas] |
| 11 |
Hb_002739_160 |
0.1211166367 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_028841_010 |
0.1212215361 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase CES101 [Jatropha curcas] |
| 13 |
Hb_030827_080 |
0.1230423022 |
- |
- |
PREDICTED: uncharacterized protein LOC105648491 [Jatropha curcas] |
| 14 |
Hb_000571_060 |
0.1244822335 |
- |
- |
PREDICTED: uncharacterized protein LOC105648112 [Jatropha curcas] |
| 15 |
Hb_033053_020 |
0.1301035653 |
- |
- |
hypothetical protein JCGZ_01204 [Jatropha curcas] |
| 16 |
Hb_004631_100 |
0.1307645058 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH96 [Jatropha curcas] |
| 17 |
Hb_038718_010 |
0.130911448 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 18 |
Hb_004324_100 |
0.1310592356 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 19 |
Hb_183209_020 |
0.1315550324 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001048_070 |
0.1328623791 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0007s14590g [Populus trichocarpa] |