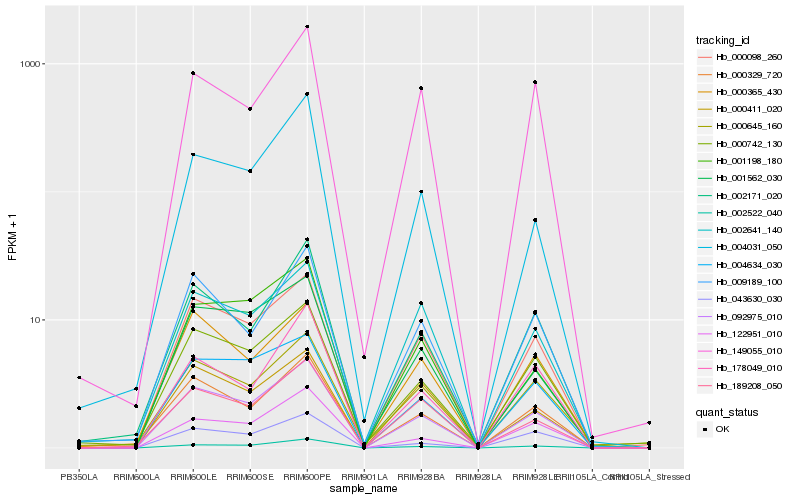
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_092975_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_04359 [Jatropha curcas] |
| 2 |
Hb_000098_260 |
0.0676531056 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 3 |
Hb_002522_040 |
0.0724770839 |
- |
- |
hypothetical protein JCGZ_04542 [Jatropha curcas] |
| 4 |
Hb_000329_720 |
0.0846806346 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0014s05500g [Populus trichocarpa] |
| 5 |
Hb_000742_130 |
0.0916232198 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g30570 [Jatropha curcas] |
| 6 |
Hb_002171_020 |
0.0960466761 |
- |
- |
protein phosphatase 2a, regulatory subunit, putative [Ricinus communis] |
| 7 |
Hb_004634_030 |
0.0972241662 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001562_030 |
0.0994040379 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH35 isoform X1 [Jatropha curcas] |
| 9 |
Hb_189208_050 |
0.1016402925 |
- |
- |
PREDICTED: uncharacterized protein LOC105642694 isoform X2 [Jatropha curcas] |
| 10 |
Hb_009189_100 |
0.1026597631 |
- |
- |
PREDICTED: patellin-6 [Jatropha curcas] |
| 11 |
Hb_122951_010 |
0.1052703023 |
- |
- |
PREDICTED: uncharacterized protein LOC105639235 [Jatropha curcas] |
| 12 |
Hb_004031_050 |
0.1086386276 |
- |
- |
sinapyl alcohol dehydrogenase-like protein [Populus tremula x Populus tremuloides] |
| 13 |
Hb_043630_030 |
0.1114850546 |
- |
- |
leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 14 |
Hb_001198_180 |
0.1141779941 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 15 |
Hb_149055_010 |
0.1147586118 |
- |
- |
cinnamate 4-hydroxylase [Leucaena leucocephala] |
| 16 |
Hb_000645_160 |
0.1152427567 |
- |
- |
protein translocase, putative [Ricinus communis] |
| 17 |
Hb_178049_010 |
0.1169162423 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000365_430 |
0.1175771813 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g08570-like [Jatropha curcas] |
| 19 |
Hb_000411_020 |
0.1187086303 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_002641_140 |
0.1199595876 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |