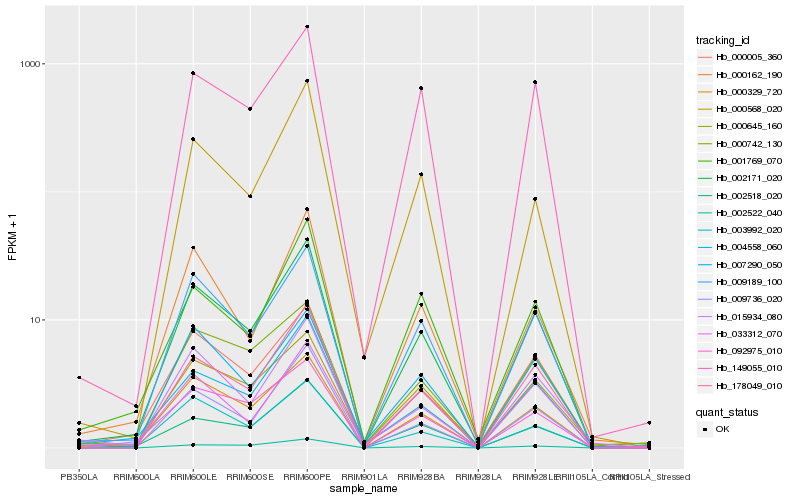
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002171_020 |
0.0 |
- |
- |
protein phosphatase 2a, regulatory subunit, putative [Ricinus communis] |
| 2 |
Hb_009189_100 |
0.0632926926 |
- |
- |
PREDICTED: patellin-6 [Jatropha curcas] |
| 3 |
Hb_000329_720 |
0.0664296049 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0014s05500g [Populus trichocarpa] |
| 4 |
Hb_178049_010 |
0.0740135619 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000005_360 |
0.0838411375 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004558_060 |
0.0843010043 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor B-4 [Jatropha curcas] |
| 7 |
Hb_015934_080 |
0.086440139 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 8 |
Hb_009736_020 |
0.0936373633 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG13 homolog [Jatropha curcas] |
| 9 |
Hb_002518_020 |
0.0958835863 |
- |
- |
PREDICTED: uncharacterized protein LOC105631164 [Jatropha curcas] |
| 10 |
Hb_092975_010 |
0.0960466761 |
- |
- |
hypothetical protein JCGZ_04359 [Jatropha curcas] |
| 11 |
Hb_149055_010 |
0.1041426961 |
- |
- |
cinnamate 4-hydroxylase [Leucaena leucocephala] |
| 12 |
Hb_000162_190 |
0.1050994615 |
- |
- |
PREDICTED: thaumatin-like protein 1b [Jatropha curcas] |
| 13 |
Hb_033312_070 |
0.1083104948 |
- |
- |
hypothetical protein POPTR_0017s02960g [Populus trichocarpa] |
| 14 |
Hb_001769_070 |
0.1091386976 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_002522_040 |
0.109784666 |
- |
- |
hypothetical protein JCGZ_04542 [Jatropha curcas] |
| 16 |
Hb_000568_020 |
0.1113587453 |
- |
- |
PREDICTED: probable aquaporin PIP2-5 [Jatropha curcas] |
| 17 |
Hb_000742_130 |
0.1116102606 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g30570 [Jatropha curcas] |
| 18 |
Hb_000645_160 |
0.1118798947 |
- |
- |
protein translocase, putative [Ricinus communis] |
| 19 |
Hb_007290_050 |
0.1124928866 |
- |
- |
PREDICTED: uncharacterized protein LOC105629472 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003992_020 |
0.1134300765 |
- |
- |
PREDICTED: phosphatidylcholine:diacylglycerol cholinephosphotransferase 1-like [Jatropha curcas] |