| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
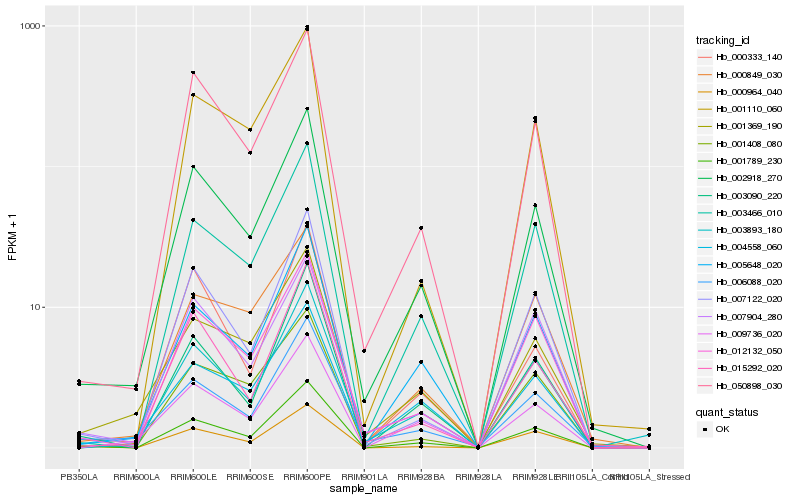
Hb_002918_270 |
0.0 |
- |
- |
PREDICTED: protein E6-like [Populus euphratica] |
| 2 |
Hb_050898_030 |
0.0655129042 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001789_230 |
0.0798077052 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005648_020 |
0.0918327044 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM1 [Jatropha curcas] |
| 5 |
Hb_015292_020 |
0.0927507777 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_003466_010 |
0.0940917118 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 2.11-like [Jatropha curcas] |
| 7 |
Hb_012132_050 |
0.0956356982 |
- |
- |
PREDICTED: aspartic proteinase PCS1 [Jatropha curcas] |
| 8 |
Hb_001369_190 |
0.0963688851 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase TDR [Jatropha curcas] |
| 9 |
Hb_009736_020 |
0.0969632239 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG13 homolog [Jatropha curcas] |
| 10 |
Hb_006088_020 |
0.0970733334 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 11 |
Hb_007122_020 |
0.09921868 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 12 |
Hb_004558_060 |
0.1001108891 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor B-4 [Jatropha curcas] |
| 13 |
Hb_003893_180 |
0.1019369469 |
- |
- |
PREDICTED: uncharacterized protein LOC105648771 [Jatropha curcas] |
| 14 |
Hb_007904_280 |
0.1068865021 |
- |
- |
PREDICTED: uncharacterized protein LOC105644987 [Jatropha curcas] |
| 15 |
Hb_003090_220 |
0.1107949487 |
- |
- |
PREDICTED: copper transporter 4 [Jatropha curcas] |
| 16 |
Hb_001408_080 |
0.1108152459 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM3 [Jatropha curcas] |
| 17 |
Hb_000849_030 |
0.1125475878 |
- |
- |
PREDICTED: fimbrin-2 [Jatropha curcas] |
| 18 |
Hb_000964_040 |
0.1143334327 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER isoform X2 [Jatropha curcas] |
| 19 |
Hb_000333_140 |
0.1143952233 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 20 |
Hb_001110_060 |
0.1156017922 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |