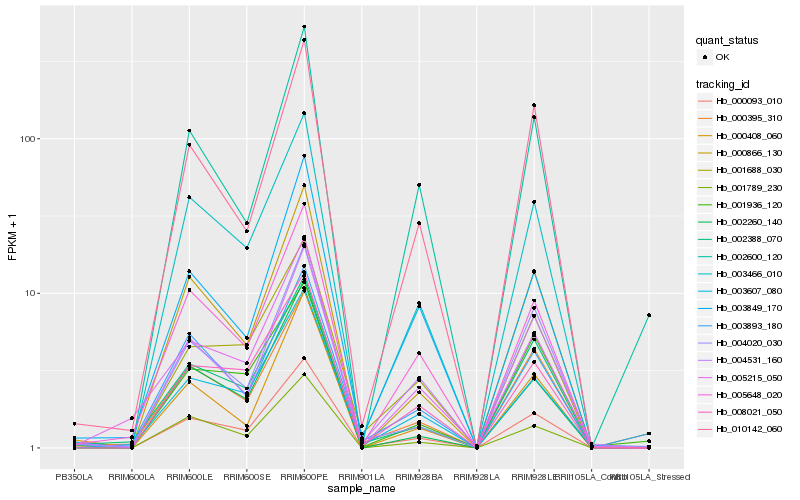
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000093_010 |
0.0 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 2 |
Hb_003607_080 |
0.0728533148 |
- |
- |
PREDICTED: putative kinase-like protein TMKL1 [Jatropha curcas] |
| 3 |
Hb_003466_010 |
0.0728606667 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 2.11-like [Jatropha curcas] |
| 4 |
Hb_005648_020 |
0.0841842946 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM1 [Jatropha curcas] |
| 5 |
Hb_008021_050 |
0.086537922 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_000395_310 |
0.0883125036 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 7 |
Hb_001688_030 |
0.0888037274 |
- |
- |
hypothetical protein JCGZ_10716 [Jatropha curcas] |
| 8 |
Hb_002388_070 |
0.0905516274 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 9 |
Hb_000866_130 |
0.0949122922 |
- |
- |
hypothetical protein JCGZ_14397 [Jatropha curcas] |
| 10 |
Hb_004531_160 |
0.0969406126 |
- |
- |
PREDICTED: transcription factor bHLH68-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_001789_230 |
0.1010981676 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002600_120 |
0.1013423179 |
- |
- |
hypothetical protein JCGZ_25652 [Jatropha curcas] |
| 13 |
Hb_003893_180 |
0.1031462618 |
- |
- |
PREDICTED: uncharacterized protein LOC105648771 [Jatropha curcas] |
| 14 |
Hb_002260_140 |
0.105471403 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000408_060 |
0.105491138 |
- |
- |
hypothetical protein POPTR_0018s11360g [Populus trichocarpa] |
| 16 |
Hb_004020_030 |
0.1074497909 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 17 |
Hb_003849_170 |
0.1082180997 |
- |
- |
hypothetical protein JCGZ_17850 [Jatropha curcas] |
| 18 |
Hb_001936_120 |
0.1089126236 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 19 |
Hb_010142_060 |
0.1103500554 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 20 |
Hb_005215_050 |
0.1104264828 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g37250 [Jatropha curcas] |