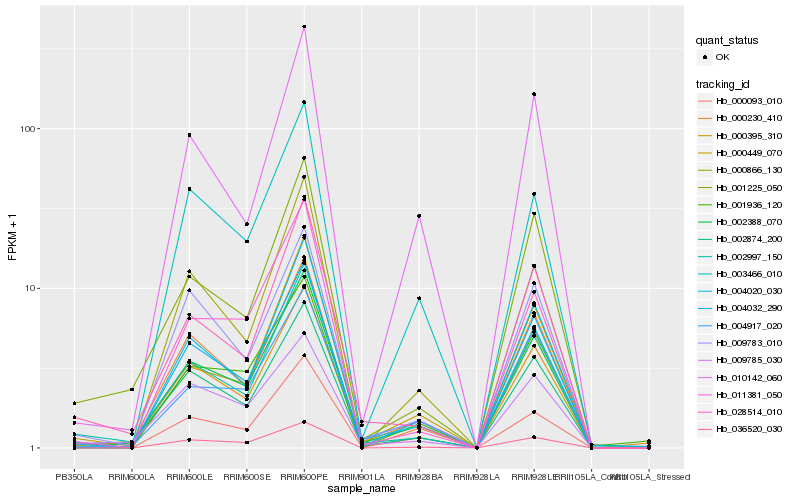
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002388_070 |
0.0 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 2 |
Hb_000395_310 |
0.0444048046 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 3 |
Hb_002874_200 |
0.0644336691 |
- |
- |
PREDICTED: uncharacterized protein LOC105109832 [Populus euphratica] |
| 4 |
Hb_002997_150 |
0.0703584096 |
- |
- |
hypothetical protein RCOM_1406750 [Ricinus communis] |
| 5 |
Hb_004032_290 |
0.0765193801 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_004020_030 |
0.0797222697 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 7 |
Hb_000230_410 |
0.0840419243 |
- |
- |
PREDICTED: uncharacterized protein LOC105631891 [Jatropha curcas] |
| 8 |
Hb_000093_010 |
0.0905516274 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 9 |
Hb_036520_030 |
0.0935007813 |
- |
- |
PREDICTED: uncharacterized protein LOC105638098 [Jatropha curcas] |
| 10 |
Hb_011381_050 |
0.0959104755 |
- |
- |
PREDICTED: protein GLUTAMINE DUMPER 5-like [Jatropha curcas] |
| 11 |
Hb_000449_070 |
0.0971653901 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_003466_010 |
0.0973565868 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 2.11-like [Jatropha curcas] |
| 13 |
Hb_004917_020 |
0.0977583028 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF2.2-like isoform X2 [Populus euphratica] |
| 14 |
Hb_001225_050 |
0.0981415258 |
transcription factor |
TF Family: LIM |
Cysteine and glycine-rich protein, putative [Ricinus communis] |
| 15 |
Hb_000866_130 |
0.0987710872 |
- |
- |
hypothetical protein JCGZ_14397 [Jatropha curcas] |
| 16 |
Hb_001936_120 |
0.0995986836 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 17 |
Hb_028514_010 |
0.0996242529 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A9 [Jatropha curcas] |
| 18 |
Hb_009783_010 |
0.1013607411 |
- |
- |
PREDICTED: sulfated surface glycoprotein 185-like [Populus euphratica] |
| 19 |
Hb_010142_060 |
0.1037471672 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 20 |
Hb_009785_030 |
0.1038528258 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor ONAC010 [Jatropha curcas] |