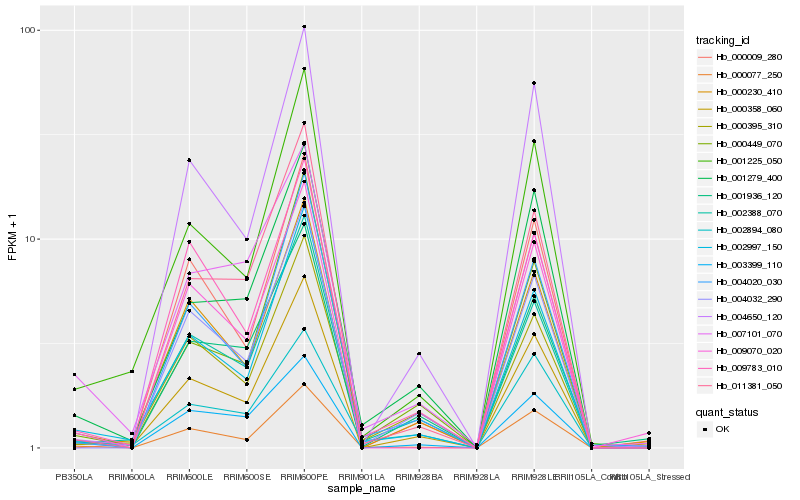
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000358_060 |
0.0 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 2 |
Hb_002997_150 |
0.0868103261 |
- |
- |
hypothetical protein RCOM_1406750 [Ricinus communis] |
| 3 |
Hb_001225_050 |
0.0930463474 |
transcription factor |
TF Family: LIM |
Cysteine and glycine-rich protein, putative [Ricinus communis] |
| 4 |
Hb_000230_410 |
0.0945742505 |
- |
- |
PREDICTED: uncharacterized protein LOC105631891 [Jatropha curcas] |
| 5 |
Hb_001936_120 |
0.0970090629 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000077_250 |
0.0996195376 |
- |
- |
hypothetical protein JCGZ_25802 [Jatropha curcas] |
| 7 |
Hb_002388_070 |
0.1063320207 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 8 |
Hb_003399_110 |
0.1095283761 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001279_400 |
0.1105457802 |
- |
- |
PREDICTED: uncharacterized protein LOC105633544 [Jatropha curcas] |
| 10 |
Hb_009070_020 |
0.1114630522 |
- |
- |
PREDICTED: uncharacterized protein LOC102612959 [Citrus sinensis] |
| 11 |
Hb_004650_120 |
0.1122682835 |
- |
- |
PREDICTED: auxin transporter-like protein 3 [Jatropha curcas] |
| 12 |
Hb_004020_030 |
0.1138845785 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 13 |
Hb_000009_280 |
0.1168736869 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 14 |
Hb_000449_070 |
0.1189565922 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_000395_310 |
0.1197809429 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 16 |
Hb_004032_290 |
0.1201150498 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_011381_050 |
0.1220245666 |
- |
- |
PREDICTED: protein GLUTAMINE DUMPER 5-like [Jatropha curcas] |
| 18 |
Hb_002894_080 |
0.1235532552 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_007101_070 |
0.1245970858 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1 [Jatropha curcas] |
| 20 |
Hb_009783_010 |
0.1260446151 |
- |
- |
PREDICTED: sulfated surface glycoprotein 185-like [Populus euphratica] |