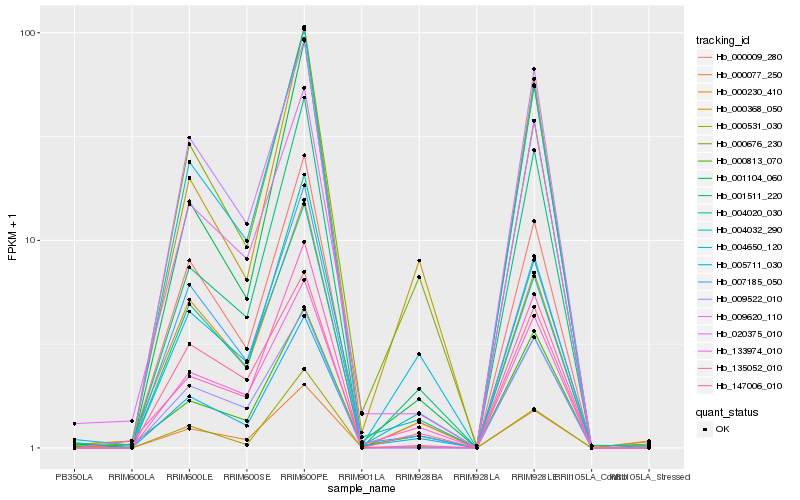
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004650_120 |
0.0 |
- |
- |
PREDICTED: auxin transporter-like protein 3 [Jatropha curcas] |
| 2 |
Hb_000009_280 |
0.061722968 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 3 |
Hb_001511_220 |
0.0675440983 |
- |
- |
hypothetical protein POPTR_0008s06990g [Populus trichocarpa] |
| 4 |
Hb_000676_230 |
0.0735241248 |
- |
- |
PREDICTED: beta-galactosidase 3 [Jatropha curcas] |
| 5 |
Hb_004032_290 |
0.0763352029 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_001104_060 |
0.0766482593 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9 [Jatropha curcas] |
| 7 |
Hb_007185_050 |
0.0775523969 |
- |
- |
hypothetical protein JCGZ_22475 [Jatropha curcas] |
| 8 |
Hb_005711_030 |
0.0775688691 |
- |
- |
hypothetical protein RCOM_0460020 [Ricinus communis] |
| 9 |
Hb_133974_010 |
0.0813694403 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 3-like isoform X2 [Populus euphratica] |
| 10 |
Hb_000077_250 |
0.0845284651 |
- |
- |
hypothetical protein JCGZ_25802 [Jatropha curcas] |
| 11 |
Hb_000230_410 |
0.0853756904 |
- |
- |
PREDICTED: uncharacterized protein LOC105631891 [Jatropha curcas] |
| 12 |
Hb_004020_030 |
0.0870019108 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 13 |
Hb_020375_010 |
0.0873634352 |
- |
- |
PREDICTED: transcription factor IBH1 [Jatropha curcas] |
| 14 |
Hb_135052_010 |
0.0902522035 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 15 |
Hb_000368_050 |
0.0942001252 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 16 |
Hb_009522_010 |
0.0960357597 |
- |
- |
hypothetical protein POPTR_0006s18790g [Populus trichocarpa] |
| 17 |
Hb_000813_070 |
0.0981000161 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_147006_010 |
0.0983991829 |
- |
- |
receptor like protein 35 [Arabidopsis thaliana] |
| 19 |
Hb_009620_110 |
0.1008175997 |
- |
- |
- |
| 20 |
Hb_000531_030 |
0.1010615653 |
- |
- |
conserved hypothetical protein [Ricinus communis] |