| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
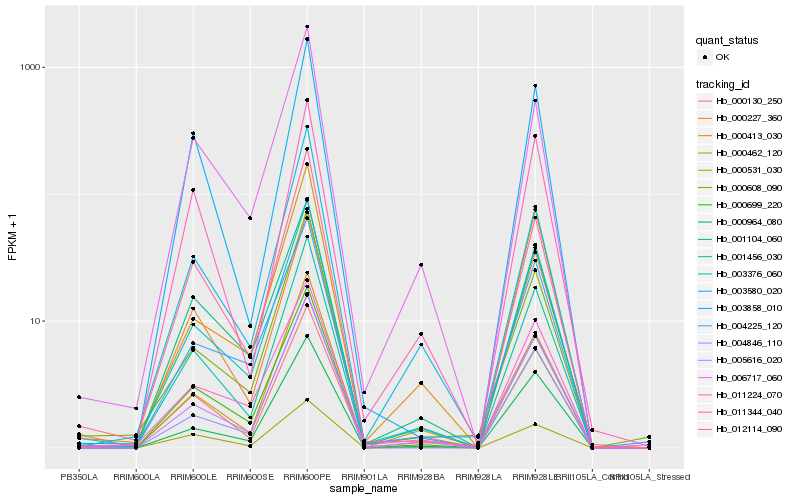
Hb_001456_030 |
0.0 |
- |
- |
PREDICTED: bifunctional pinoresinol-lariciresinol reductase 2 [Vitis vinifera] |
| 2 |
Hb_005616_020 |
0.044602086 |
- |
- |
kinase, putative [Ricinus communis] |
| 3 |
Hb_003376_060 |
0.0452988057 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000227_360 |
0.0693203992 |
- |
- |
PREDICTED: mavicyanin [Jatropha curcas] |
| 5 |
Hb_000462_120 |
0.0708818861 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105642310 [Jatropha curcas] |
| 6 |
Hb_001104_060 |
0.0737100862 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9 [Jatropha curcas] |
| 7 |
Hb_000964_080 |
0.074503299 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Jatropha curcas] |
| 8 |
Hb_000413_030 |
0.0749967246 |
- |
- |
PREDICTED: NAD(P)H dehydrogenase (quinone) FQR1-like [Jatropha curcas] |
| 9 |
Hb_012114_090 |
0.0759594005 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |
| 10 |
Hb_000699_220 |
0.0772616181 |
- |
- |
PREDICTED: phospholipid-metabolizing enzyme A-C1-like [Jatropha curcas] |
| 11 |
Hb_011224_070 |
0.0823422322 |
- |
- |
PREDICTED: solute carrier family 35 member E3 [Jatropha curcas] |
| 12 |
Hb_000531_030 |
0.0829314376 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_006717_060 |
0.0834213632 |
- |
- |
PREDICTED: peroxidase 42 [Jatropha curcas] |
| 14 |
Hb_000608_090 |
0.0847791749 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000130_250 |
0.0887622524 |
- |
- |
PREDICTED: protein trichome birefringence-like 31 [Jatropha curcas] |
| 16 |
Hb_003580_020 |
0.0920594909 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003858_010 |
0.0938364015 |
- |
- |
xyloglucan endotransglucosylase/hydrolase [Gossypium hirsutum] |
| 18 |
Hb_004225_120 |
0.0949636686 |
- |
- |
PREDICTED: uncharacterized protein LOC105628059 [Jatropha curcas] |
| 19 |
Hb_011344_040 |
0.0964088079 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 20 |
Hb_004846_110 |
0.0966773014 |
- |
- |
hypothetical protein JCGZ_04591 [Jatropha curcas] |