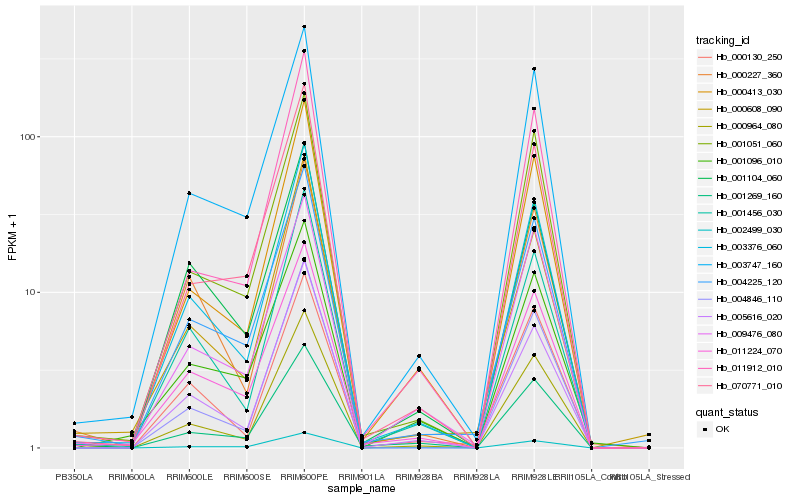
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003376_060 |
0.0 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001456_030 |
0.0452988057 |
- |
- |
PREDICTED: bifunctional pinoresinol-lariciresinol reductase 2 [Vitis vinifera] |
| 3 |
Hb_011224_070 |
0.0517038722 |
- |
- |
PREDICTED: solute carrier family 35 member E3 [Jatropha curcas] |
| 4 |
Hb_005616_020 |
0.0554971307 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_000964_080 |
0.0563323738 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Jatropha curcas] |
| 6 |
Hb_000413_030 |
0.0571228838 |
- |
- |
PREDICTED: NAD(P)H dehydrogenase (quinone) FQR1-like [Jatropha curcas] |
| 7 |
Hb_003747_160 |
0.0616275382 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004225_120 |
0.0627194943 |
- |
- |
PREDICTED: uncharacterized protein LOC105628059 [Jatropha curcas] |
| 9 |
Hb_001051_060 |
0.0712893873 |
- |
- |
UDP-glucuronic acid/UDP-N-acetylgalactosamine transporter, putative [Ricinus communis] |
| 10 |
Hb_001104_060 |
0.0722801413 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9 [Jatropha curcas] |
| 11 |
Hb_004846_110 |
0.0744148621 |
- |
- |
hypothetical protein JCGZ_04591 [Jatropha curcas] |
| 12 |
Hb_000227_360 |
0.0752528621 |
- |
- |
PREDICTED: mavicyanin [Jatropha curcas] |
| 13 |
Hb_070771_010 |
0.0755050924 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g06890 [Jatropha curcas] |
| 14 |
Hb_001269_160 |
0.0783474266 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_011912_010 |
0.0785173704 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 16 |
Hb_000130_250 |
0.0826281385 |
- |
- |
PREDICTED: protein trichome birefringence-like 31 [Jatropha curcas] |
| 17 |
Hb_000608_090 |
0.0830188149 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002499_030 |
0.0832243729 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 19 |
Hb_009476_080 |
0.0837294919 |
- |
- |
BnaA03g18920D [Brassica napus] |
| 20 |
Hb_001096_010 |
0.0859511511 |
- |
- |
PREDICTED: uncharacterized protein LOC105650448 [Jatropha curcas] |