| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
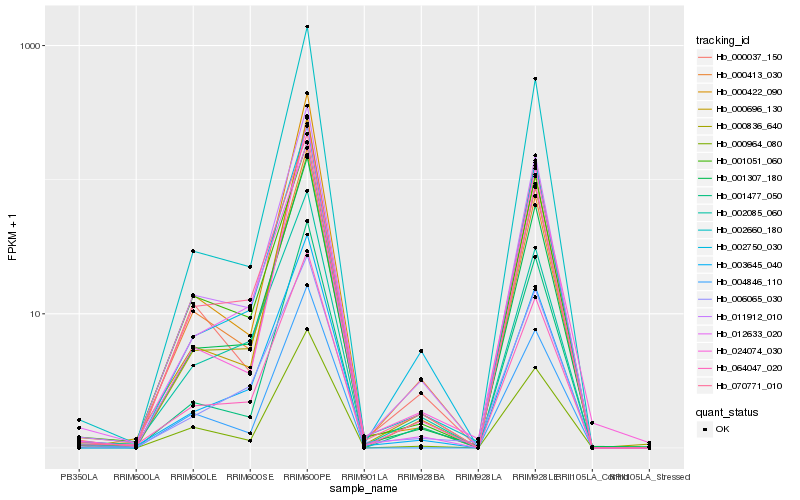
Hb_001307_180 |
0.0 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 4 [UDP-forming] isoform X1 [Jatropha curcas] |
| 2 |
Hb_011912_010 |
0.0283101239 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 3 |
Hb_003645_040 |
0.0479213944 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0003s13190g [Populus trichocarpa] |
| 4 |
Hb_012633_020 |
0.0480273494 |
- |
- |
laccase, putative [Ricinus communis] |
| 5 |
Hb_070771_010 |
0.0489199647 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g06890 [Jatropha curcas] |
| 6 |
Hb_002660_180 |
0.0528514794 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000836_640 |
0.0579820614 |
- |
- |
PREDICTED: uncharacterized protein LOC105644242 [Jatropha curcas] |
| 8 |
Hb_004846_110 |
0.0626883468 |
- |
- |
hypothetical protein JCGZ_04591 [Jatropha curcas] |
| 9 |
Hb_000413_030 |
0.0629739538 |
- |
- |
PREDICTED: NAD(P)H dehydrogenase (quinone) FQR1-like [Jatropha curcas] |
| 10 |
Hb_000037_150 |
0.0655507207 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 11 |
Hb_002750_030 |
0.0678078133 |
- |
- |
PREDICTED: protein IRX15-LIKE-like [Populus euphratica] |
| 12 |
Hb_002085_060 |
0.0704315867 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_024074_030 |
0.0705094567 |
- |
- |
hypothetical protein CISIN_1g043166mg [Citrus sinensis] |
| 14 |
Hb_064047_020 |
0.0706425557 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0003s13190g [Populus trichocarpa] |
| 15 |
Hb_001477_050 |
0.0718469476 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_001051_060 |
0.0734615471 |
- |
- |
UDP-glucuronic acid/UDP-N-acetylgalactosamine transporter, putative [Ricinus communis] |
| 17 |
Hb_000964_080 |
0.0745972917 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Jatropha curcas] |
| 18 |
Hb_000696_130 |
0.0748573776 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 19 |
Hb_000422_090 |
0.0779507007 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_006065_030 |
0.0799124911 |
- |
- |
PREDICTED: filament-like plant protein 7 isoform X1 [Jatropha curcas] |