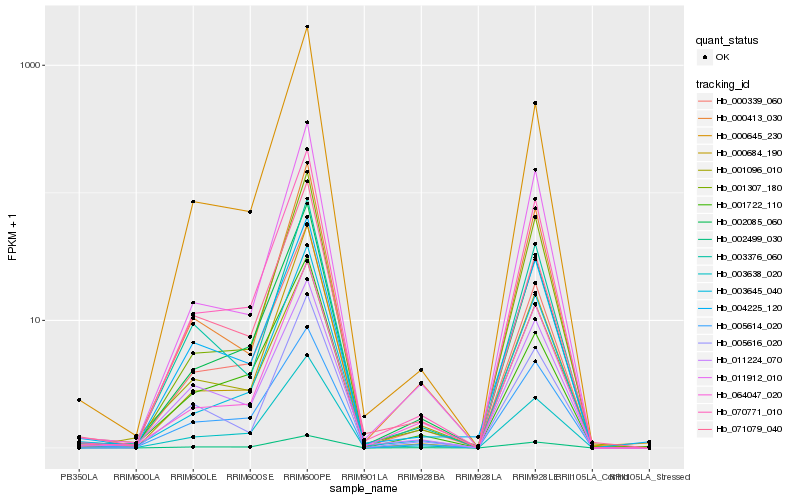
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000339_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003638_020 |
0.0273385463 |
- |
- |
PREDICTED: uncharacterized protein LOC105645807 [Jatropha curcas] |
| 3 |
Hb_002085_060 |
0.0317683322 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_070771_010 |
0.0470586639 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g06890 [Jatropha curcas] |
| 5 |
Hb_002499_030 |
0.0667697547 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 6 |
Hb_071079_040 |
0.0696067509 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET3 [Jatropha curcas] |
| 7 |
Hb_000684_190 |
0.0729662332 |
- |
- |
beta-1,3-glucuronyltransferase, putative [Ricinus communis] |
| 8 |
Hb_000645_230 |
0.0733809599 |
- |
- |
PREDICTED: chitinase-like protein 2 [Jatropha curcas] |
| 9 |
Hb_003645_040 |
0.0737586376 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0003s13190g [Populus trichocarpa] |
| 10 |
Hb_005614_020 |
0.0765968395 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X1 [Jatropha curcas] |
| 11 |
Hb_064047_020 |
0.0790332057 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0003s13190g [Populus trichocarpa] |
| 12 |
Hb_011912_010 |
0.0831562986 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 13 |
Hb_001722_110 |
0.0841237332 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 14 |
Hb_000413_030 |
0.085570358 |
- |
- |
PREDICTED: NAD(P)H dehydrogenase (quinone) FQR1-like [Jatropha curcas] |
| 15 |
Hb_004225_120 |
0.0858562542 |
- |
- |
PREDICTED: uncharacterized protein LOC105628059 [Jatropha curcas] |
| 16 |
Hb_011224_070 |
0.0874491425 |
- |
- |
PREDICTED: solute carrier family 35 member E3 [Jatropha curcas] |
| 17 |
Hb_001307_180 |
0.0878605384 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 4 [UDP-forming] isoform X1 [Jatropha curcas] |
| 18 |
Hb_001096_010 |
0.0879800016 |
- |
- |
PREDICTED: uncharacterized protein LOC105650448 [Jatropha curcas] |
| 19 |
Hb_005616_020 |
0.089874399 |
- |
- |
kinase, putative [Ricinus communis] |
| 20 |
Hb_003376_060 |
0.0922786045 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9 isoform X1 [Jatropha curcas] |