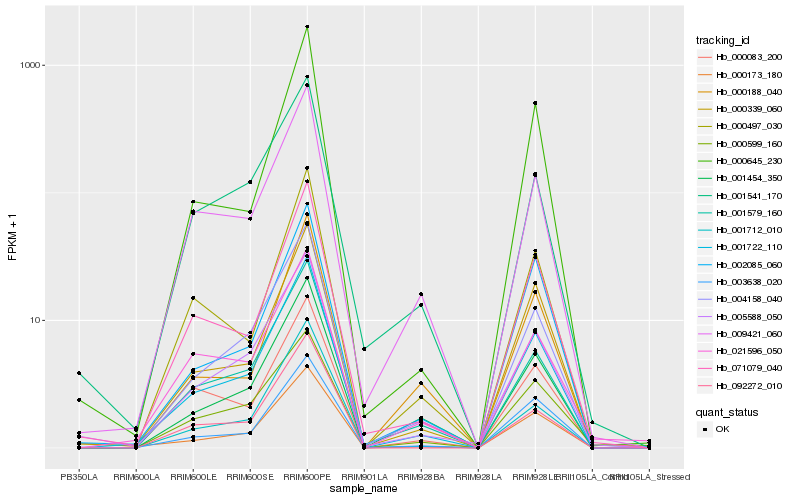
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001722_110 |
0.0 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_004158_040 |
0.060574548 |
- |
- |
PREDICTED: WAT1-related protein At2g37460-like [Jatropha curcas] |
| 3 |
Hb_003638_020 |
0.078167662 |
- |
- |
PREDICTED: uncharacterized protein LOC105645807 [Jatropha curcas] |
| 4 |
Hb_001579_160 |
0.078214407 |
- |
- |
PREDICTED: kininogen-1-like [Jatropha curcas] |
| 5 |
Hb_001454_350 |
0.0789634175 |
- |
- |
PREDICTED: uncharacterized protein LOC105643449 [Jatropha curcas] |
| 6 |
Hb_005588_050 |
0.0836152978 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000339_060 |
0.0841237332 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 8 |
Hb_009421_060 |
0.0846124832 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 9 |
Hb_071079_040 |
0.0853859824 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET3 [Jatropha curcas] |
| 10 |
Hb_001712_010 |
0.0963530072 |
- |
- |
PREDICTED: phosphate transporter PHO1 homolog 1 [Jatropha curcas] |
| 11 |
Hb_092272_010 |
0.0973300079 |
- |
- |
- |
| 12 |
Hb_000497_030 |
0.0981516141 |
- |
- |
- |
| 13 |
Hb_021596_050 |
0.0993848854 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000645_230 |
0.1001883296 |
- |
- |
PREDICTED: chitinase-like protein 2 [Jatropha curcas] |
| 15 |
Hb_000083_200 |
0.1013588476 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002085_060 |
0.103534713 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000173_180 |
0.1083631852 |
- |
- |
PREDICTED: adenylate isopentenyltransferase 5, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001541_170 |
0.1084485477 |
- |
- |
hypothetical protein JCGZ_17850 [Jatropha curcas] |
| 19 |
Hb_000599_160 |
0.1091954545 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 21/22 [Jatropha curcas] |
| 20 |
Hb_000188_040 |
0.1095875461 |
- |
- |
PREDICTED: uncharacterized protein LOC105629522 [Jatropha curcas] |