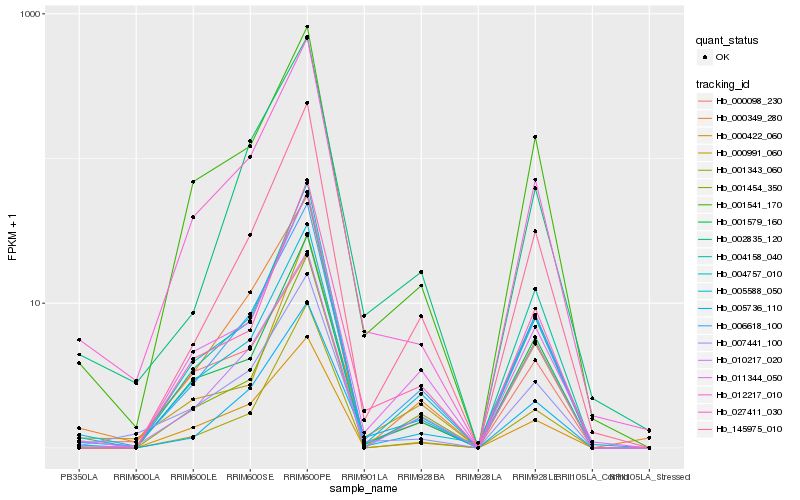
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006618_100 |
0.0 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus tremula x Populus tremuloides] |
| 2 |
Hb_010217_020 |
0.0599712843 |
- |
- |
cinnamate 4-hydroxylase [Leucaena leucocephala] |
| 3 |
Hb_145975_010 |
0.0685252276 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 4 |
Hb_000349_280 |
0.0745228585 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004757_010 |
0.0808872249 |
- |
- |
PREDICTED: uncharacterized protein LOC105121200 [Populus euphratica] |
| 6 |
Hb_004158_040 |
0.0818233308 |
- |
- |
PREDICTED: WAT1-related protein At2g37460-like [Jatropha curcas] |
| 7 |
Hb_001579_160 |
0.0839223058 |
- |
- |
PREDICTED: kininogen-1-like [Jatropha curcas] |
| 8 |
Hb_001541_170 |
0.0851324684 |
- |
- |
hypothetical protein JCGZ_17850 [Jatropha curcas] |
| 9 |
Hb_011344_050 |
0.0874283826 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 10 |
Hb_001454_350 |
0.0935177972 |
- |
- |
PREDICTED: uncharacterized protein LOC105643449 [Jatropha curcas] |
| 11 |
Hb_005736_110 |
0.0972186427 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus tremula x Populus tremuloides] |
| 12 |
Hb_027411_030 |
0.1007697695 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 13 |
Hb_000422_060 |
0.1024082115 |
transcription factor |
TF Family: OFP |
hypothetical protein RCOM_1282480 [Ricinus communis] |
| 14 |
Hb_005588_050 |
0.1029162798 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_007441_100 |
0.1035916002 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 12 [Populus euphratica] |
| 16 |
Hb_012217_010 |
0.1044415705 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A9 [Jatropha curcas] |
| 17 |
Hb_000098_230 |
0.1075160335 |
- |
- |
Potassium channel AKT2/3, putative [Ricinus communis] |
| 18 |
Hb_002835_120 |
0.1110067201 |
- |
- |
PREDICTED: uncharacterized protein LOC105640304 [Jatropha curcas] |
| 19 |
Hb_001343_060 |
0.114513181 |
- |
- |
Cytochrome P450 [Theobroma cacao] |
| 20 |
Hb_000991_060 |
0.1161720978 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |