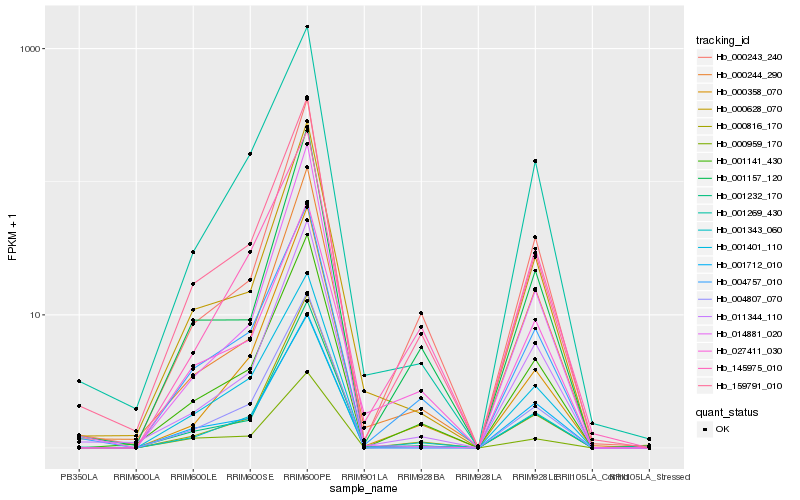
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001343_060 |
0.0 |
- |
- |
Cytochrome P450 [Theobroma cacao] |
| 2 |
Hb_001141_430 |
0.0447586072 |
- |
- |
hypothetical protein POPTR_0008s15160g [Populus trichocarpa] |
| 3 |
Hb_159791_010 |
0.0598403307 |
- |
- |
PREDICTED: cytochrome P450 84A1 [Jatropha curcas] |
| 4 |
Hb_004757_010 |
0.0602071161 |
- |
- |
PREDICTED: uncharacterized protein LOC105121200 [Populus euphratica] |
| 5 |
Hb_011344_110 |
0.0629422137 |
transcription factor |
TF Family: MYB |
PREDICTED: protein ODORANT1 [Jatropha curcas] |
| 6 |
Hb_001269_430 |
0.0640897602 |
- |
- |
hypothetical protein JCGZ_04286 [Jatropha curcas] |
| 7 |
Hb_000816_170 |
0.0693132653 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 15 [Jatropha curcas] |
| 8 |
Hb_001712_010 |
0.0725622352 |
- |
- |
PREDICTED: phosphate transporter PHO1 homolog 1 [Jatropha curcas] |
| 9 |
Hb_004807_070 |
0.0747511355 |
- |
- |
hypothetical protein POPTR_0004s17330g, partial [Populus trichocarpa] |
| 10 |
Hb_000243_240 |
0.0757901411 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 11 |
Hb_000244_290 |
0.0793199305 |
- |
- |
PREDICTED: UDP-glucuronate:xylan alpha-glucuronosyltransferase 1 [Jatropha curcas] |
| 12 |
Hb_001232_170 |
0.0814347737 |
- |
- |
sulfate transporter, putative [Ricinus communis] |
| 13 |
Hb_145975_010 |
0.0863068913 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 14 |
Hb_000628_070 |
0.086590943 |
- |
- |
- |
| 15 |
Hb_001401_110 |
0.0908786051 |
- |
- |
hypothetical protein JCGZ_04272 [Jatropha curcas] |
| 16 |
Hb_001157_120 |
0.0913048092 |
- |
- |
PREDICTED: dirigent protein 19-like [Jatropha curcas] |
| 17 |
Hb_000358_070 |
0.0946854246 |
- |
- |
hypothetical protein POPTR_0003s01410g [Populus trichocarpa] |
| 18 |
Hb_027411_030 |
0.0971547194 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 19 |
Hb_000959_170 |
0.0973386901 |
- |
- |
Cell division protein kinase 7, putative [Ricinus communis] |
| 20 |
Hb_014881_020 |
0.0978016815 |
- |
- |
Protein P21, putative [Ricinus communis] |