| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
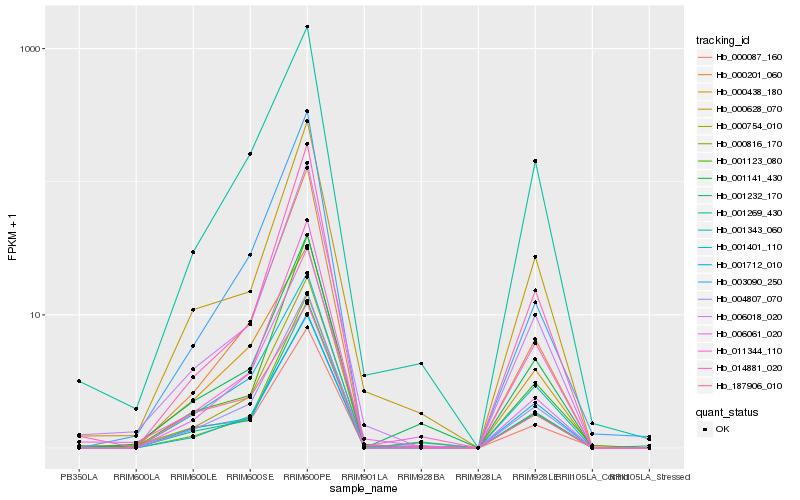
Hb_004807_070 |
0.0 |
- |
- |
hypothetical protein POPTR_0004s17330g, partial [Populus trichocarpa] |
| 2 |
Hb_001401_110 |
0.0536472379 |
- |
- |
hypothetical protein JCGZ_04272 [Jatropha curcas] |
| 3 |
Hb_001269_430 |
0.067893003 |
- |
- |
hypothetical protein JCGZ_04286 [Jatropha curcas] |
| 4 |
Hb_000201_060 |
0.0697324368 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X2 [Populus euphratica] |
| 5 |
Hb_000754_010 |
0.0700200957 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_006018_020 |
0.0741674292 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001343_060 |
0.0747511355 |
- |
- |
Cytochrome P450 [Theobroma cacao] |
| 8 |
Hb_006061_020 |
0.0749587907 |
transcription factor |
TF Family: bZIP |
bZIP family protein [Populus trichocarpa] |
| 9 |
Hb_001232_170 |
0.0790975824 |
- |
- |
sulfate transporter, putative [Ricinus communis] |
| 10 |
Hb_001712_010 |
0.0818397756 |
- |
- |
PREDICTED: phosphate transporter PHO1 homolog 1 [Jatropha curcas] |
| 11 |
Hb_000438_180 |
0.0820975395 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 12 |
Hb_001141_430 |
0.0833893695 |
- |
- |
hypothetical protein POPTR_0008s15160g [Populus trichocarpa] |
| 13 |
Hb_014881_020 |
0.0835408712 |
- |
- |
Protein P21, putative [Ricinus communis] |
| 14 |
Hb_000816_170 |
0.0837263226 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 15 [Jatropha curcas] |
| 15 |
Hb_003090_250 |
0.0843783965 |
- |
- |
Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein isoform 1 [Theobroma cacao] |
| 16 |
Hb_011344_110 |
0.0874865519 |
transcription factor |
TF Family: MYB |
PREDICTED: protein ODORANT1 [Jatropha curcas] |
| 17 |
Hb_000628_070 |
0.0887013189 |
- |
- |
- |
| 18 |
Hb_001123_080 |
0.0902768028 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000087_160 |
0.090928132 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 20 |
Hb_187906_010 |
0.0923571754 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA2-like [Populus euphratica] |