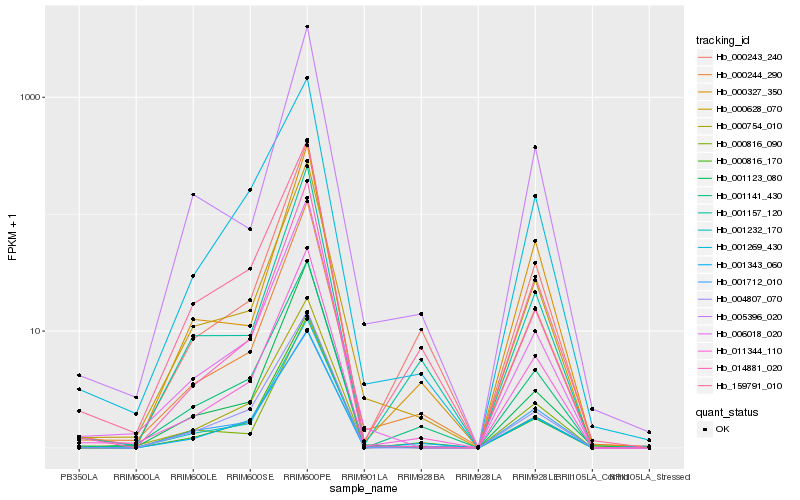
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011344_110 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: protein ODORANT1 [Jatropha curcas] |
| 2 |
Hb_000244_290 |
0.0419974056 |
- |
- |
PREDICTED: UDP-glucuronate:xylan alpha-glucuronosyltransferase 1 [Jatropha curcas] |
| 3 |
Hb_001343_060 |
0.0629422137 |
- |
- |
Cytochrome P450 [Theobroma cacao] |
| 4 |
Hb_000628_070 |
0.0637521082 |
- |
- |
- |
| 5 |
Hb_014881_020 |
0.0664230338 |
- |
- |
Protein P21, putative [Ricinus communis] |
| 6 |
Hb_001232_170 |
0.0682700808 |
- |
- |
sulfate transporter, putative [Ricinus communis] |
| 7 |
Hb_006018_020 |
0.0707833377 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001141_430 |
0.0718088441 |
- |
- |
hypothetical protein POPTR_0008s15160g [Populus trichocarpa] |
| 9 |
Hb_000816_170 |
0.076227826 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 15 [Jatropha curcas] |
| 10 |
Hb_000243_240 |
0.0766059678 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 11 |
Hb_001712_010 |
0.077275603 |
- |
- |
PREDICTED: phosphate transporter PHO1 homolog 1 [Jatropha curcas] |
| 12 |
Hb_001269_430 |
0.0803290053 |
- |
- |
hypothetical protein JCGZ_04286 [Jatropha curcas] |
| 13 |
Hb_000816_090 |
0.0850038478 |
transcription factor |
TF Family: TRAF |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_001157_120 |
0.0873512489 |
- |
- |
PREDICTED: dirigent protein 19-like [Jatropha curcas] |
| 15 |
Hb_004807_070 |
0.0874865519 |
- |
- |
hypothetical protein POPTR_0004s17330g, partial [Populus trichocarpa] |
| 16 |
Hb_159791_010 |
0.0909541194 |
- |
- |
PREDICTED: cytochrome P450 84A1 [Jatropha curcas] |
| 17 |
Hb_001123_080 |
0.0911387249 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000327_350 |
0.0935131739 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 21-like [Jatropha curcas] |
| 19 |
Hb_005396_020 |
0.0955857702 |
- |
- |
hypothetical protein CICLE_v10033278mg [Citrus clementina] |
| 20 |
Hb_000754_010 |
0.1007843355 |
- |
- |
unnamed protein product [Vitis vinifera] |