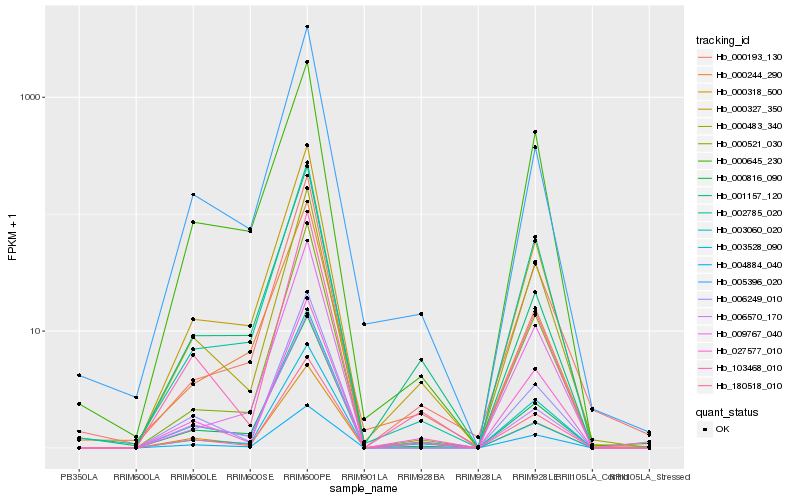
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000327_350 |
0.0 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 21-like [Jatropha curcas] |
| 2 |
Hb_000816_090 |
0.0459862724 |
transcription factor |
TF Family: TRAF |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_003060_020 |
0.0514264382 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 4 |
Hb_000521_030 |
0.0695354118 |
- |
- |
PREDICTED: uncharacterized protein LOC105650350 [Jatropha curcas] |
| 5 |
Hb_003528_090 |
0.0714913858 |
- |
- |
Uncharacterized protein TCM_037169 [Theobroma cacao] |
| 6 |
Hb_000318_500 |
0.0736260521 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000244_290 |
0.0738293691 |
- |
- |
PREDICTED: UDP-glucuronate:xylan alpha-glucuronosyltransferase 1 [Jatropha curcas] |
| 8 |
Hb_002785_020 |
0.0757074608 |
- |
- |
PREDICTED: laccase-4-like [Jatropha curcas] |
| 9 |
Hb_005396_020 |
0.0761753626 |
- |
- |
hypothetical protein CICLE_v10033278mg [Citrus clementina] |
| 10 |
Hb_180518_010 |
0.0785743489 |
- |
- |
PREDICTED: receptor-like protein 2 [Jatropha curcas] |
| 11 |
Hb_000193_130 |
0.0794147023 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 12 |
Hb_103468_010 |
0.0822517302 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 13 |
Hb_009767_040 |
0.0838957936 |
- |
- |
PREDICTED: uncharacterized protein LOC105127022 [Populus euphratica] |
| 14 |
Hb_027577_010 |
0.0847717983 |
- |
- |
PREDICTED: uncharacterized protein LOC104779928 [Camelina sativa] |
| 15 |
Hb_000645_230 |
0.0905979852 |
- |
- |
PREDICTED: chitinase-like protein 2 [Jatropha curcas] |
| 16 |
Hb_006570_170 |
0.0913993677 |
- |
- |
Gibberellin-regulated protein 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_006249_010 |
0.0919342372 |
- |
- |
hypothetical protein JCGZ_13152 [Jatropha curcas] |
| 18 |
Hb_000483_340 |
0.0921066804 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004884_040 |
0.0923401887 |
- |
- |
hypothetical protein POPTR_0016s12910g [Populus trichocarpa] |
| 20 |
Hb_001157_120 |
0.0925463116 |
- |
- |
PREDICTED: dirigent protein 19-like [Jatropha curcas] |