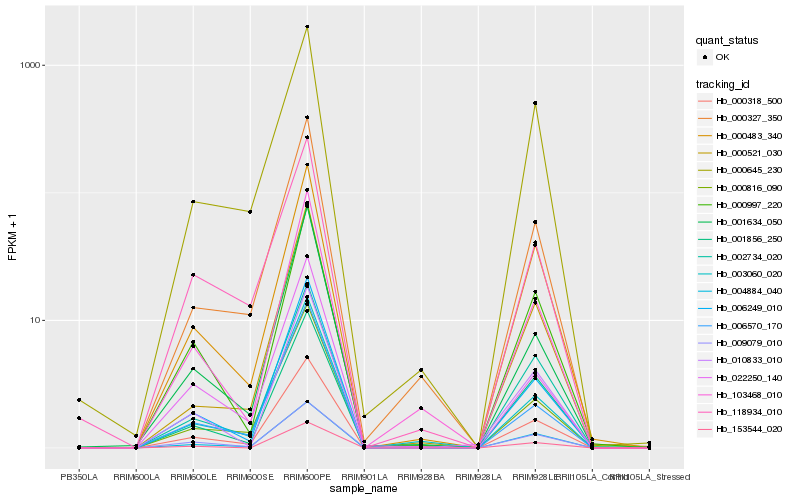
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000318_500 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004884_040 |
0.0530069713 |
- |
- |
hypothetical protein POPTR_0016s12910g [Populus trichocarpa] |
| 3 |
Hb_006249_010 |
0.0550231993 |
- |
- |
hypothetical protein JCGZ_13152 [Jatropha curcas] |
| 4 |
Hb_000483_340 |
0.0644738755 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002734_020 |
0.0659723755 |
- |
- |
PREDICTED: uncharacterized protein LOC105631540 [Jatropha curcas] |
| 6 |
Hb_000816_090 |
0.0695971423 |
transcription factor |
TF Family: TRAF |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_000327_350 |
0.0736260521 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 21-like [Jatropha curcas] |
| 8 |
Hb_000997_220 |
0.0736940915 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA14-like [Jatropha curcas] |
| 9 |
Hb_006570_170 |
0.0745177637 |
- |
- |
Gibberellin-regulated protein 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_001856_250 |
0.0768067554 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Jatropha curcas] |
| 11 |
Hb_010833_010 |
0.0804209373 |
- |
- |
hypothetical protein JCGZ_05701 [Jatropha curcas] |
| 12 |
Hb_103468_010 |
0.081344777 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 13 |
Hb_003060_020 |
0.0813458244 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 14 |
Hb_009079_010 |
0.0822445163 |
- |
- |
JHL18I08.8 [Jatropha curcas] |
| 15 |
Hb_153544_020 |
0.0826296874 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP12 [Jatropha curcas] |
| 16 |
Hb_022250_140 |
0.0831425364 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 17 |
Hb_118934_010 |
0.0847125596 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 18 |
Hb_001634_050 |
0.0848630828 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 12 [Jatropha curcas] |
| 19 |
Hb_000521_030 |
0.0864258607 |
- |
- |
PREDICTED: uncharacterized protein LOC105650350 [Jatropha curcas] |
| 20 |
Hb_000645_230 |
0.0885422118 |
- |
- |
PREDICTED: chitinase-like protein 2 [Jatropha curcas] |