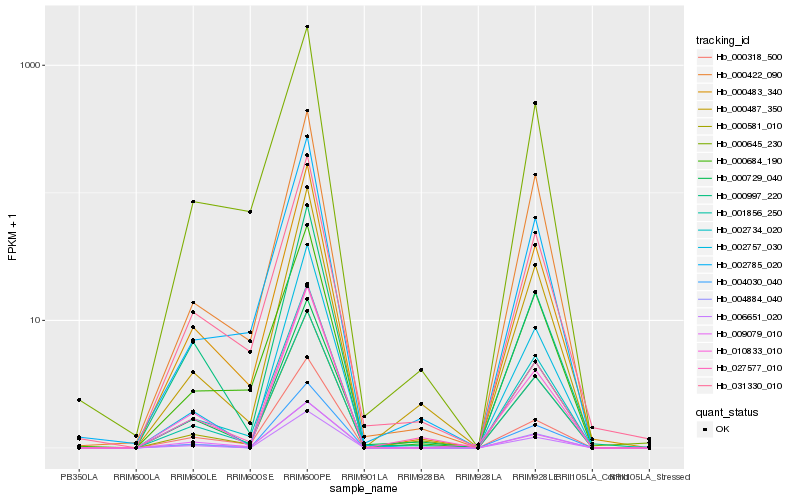
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000483_340 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001856_250 |
0.0290348336 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Jatropha curcas] |
| 3 |
Hb_004884_040 |
0.0324001567 |
- |
- |
hypothetical protein POPTR_0016s12910g [Populus trichocarpa] |
| 4 |
Hb_002734_020 |
0.0331217096 |
- |
- |
PREDICTED: uncharacterized protein LOC105631540 [Jatropha curcas] |
| 5 |
Hb_031330_010 |
0.0564855711 |
- |
- |
beta-1,3-glucuronyltransferase, putative [Ricinus communis] |
| 6 |
Hb_000997_220 |
0.0616322871 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA14-like [Jatropha curcas] |
| 7 |
Hb_000645_230 |
0.0642686925 |
- |
- |
PREDICTED: chitinase-like protein 2 [Jatropha curcas] |
| 8 |
Hb_000318_500 |
0.0644738755 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_027577_010 |
0.0658817075 |
- |
- |
PREDICTED: uncharacterized protein LOC104779928 [Camelina sativa] |
| 10 |
Hb_000422_090 |
0.0706722853 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002785_020 |
0.0718916283 |
- |
- |
PREDICTED: laccase-4-like [Jatropha curcas] |
| 12 |
Hb_000684_190 |
0.0725999695 |
- |
- |
beta-1,3-glucuronyltransferase, putative [Ricinus communis] |
| 13 |
Hb_000729_040 |
0.0739512795 |
- |
- |
xyloglucan endotransglucosylase/hydrolase protein 10 precursor [Populus trichocarpa] |
| 14 |
Hb_006651_020 |
0.0749758686 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 15 |
Hb_000487_350 |
0.0750418907 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000581_010 |
0.0787467002 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.6-like [Jatropha curcas] |
| 17 |
Hb_004030_040 |
0.0809375976 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 2.8 [Jatropha curcas] |
| 18 |
Hb_002757_030 |
0.0837375663 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_010833_010 |
0.083842948 |
- |
- |
hypothetical protein JCGZ_05701 [Jatropha curcas] |
| 20 |
Hb_009079_010 |
0.0845460441 |
- |
- |
JHL18I08.8 [Jatropha curcas] |