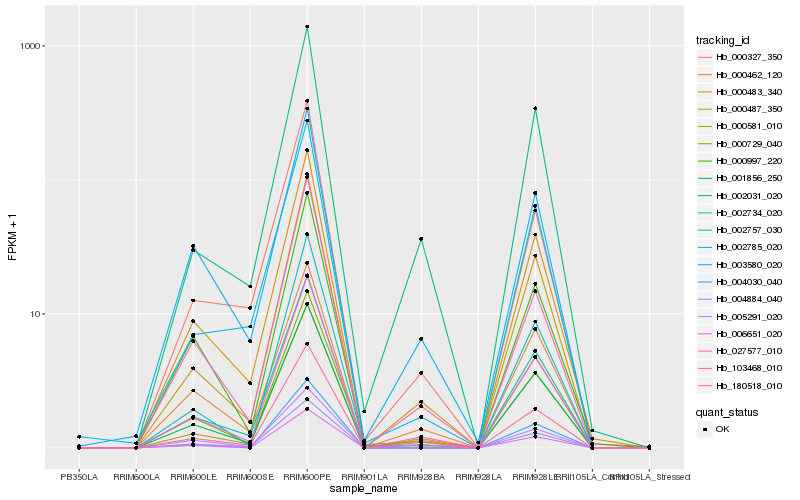
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027577_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104779928 [Camelina sativa] |
| 2 |
Hb_180518_010 |
0.0336564482 |
- |
- |
PREDICTED: receptor-like protein 2 [Jatropha curcas] |
| 3 |
Hb_000487_350 |
0.0358139765 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000581_010 |
0.0399640441 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.6-like [Jatropha curcas] |
| 5 |
Hb_001856_250 |
0.0584302867 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Jatropha curcas] |
| 6 |
Hb_000729_040 |
0.0642832329 |
- |
- |
xyloglucan endotransglucosylase/hydrolase protein 10 precursor [Populus trichocarpa] |
| 7 |
Hb_000483_340 |
0.0658817075 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002031_020 |
0.0671470298 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 9 |
Hb_103468_010 |
0.0701448875 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_002734_020 |
0.0743549274 |
- |
- |
PREDICTED: uncharacterized protein LOC105631540 [Jatropha curcas] |
| 11 |
Hb_000462_120 |
0.0774276176 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105642310 [Jatropha curcas] |
| 12 |
Hb_004884_040 |
0.0804614929 |
- |
- |
hypothetical protein POPTR_0016s12910g [Populus trichocarpa] |
| 13 |
Hb_005291_020 |
0.0806492366 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000997_220 |
0.0846330628 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA14-like [Jatropha curcas] |
| 15 |
Hb_000327_350 |
0.0847717983 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 21-like [Jatropha curcas] |
| 16 |
Hb_006651_020 |
0.0853159364 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 17 |
Hb_002757_030 |
0.0856388714 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_003580_020 |
0.086700777 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004030_040 |
0.0868269029 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 2.8 [Jatropha curcas] |
| 20 |
Hb_002785_020 |
0.087131482 |
- |
- |
PREDICTED: laccase-4-like [Jatropha curcas] |