| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
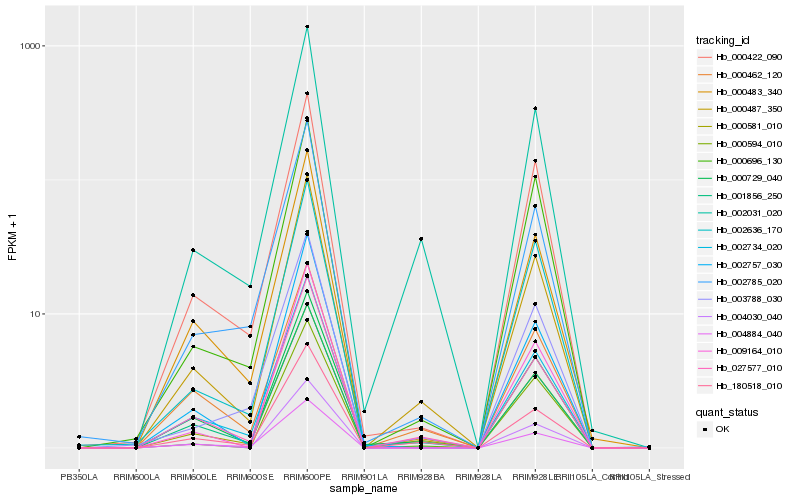
Hb_000581_010 |
0.0 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.6-like [Jatropha curcas] |
| 2 |
Hb_000487_350 |
0.0085917728 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_027577_010 |
0.0399640441 |
- |
- |
PREDICTED: uncharacterized protein LOC104779928 [Camelina sativa] |
| 4 |
Hb_002031_020 |
0.0446549208 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 5 |
Hb_180518_010 |
0.0533137627 |
- |
- |
PREDICTED: receptor-like protein 2 [Jatropha curcas] |
| 6 |
Hb_000729_040 |
0.0653648402 |
- |
- |
xyloglucan endotransglucosylase/hydrolase protein 10 precursor [Populus trichocarpa] |
| 7 |
Hb_001856_250 |
0.0675682067 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Jatropha curcas] |
| 8 |
Hb_000483_340 |
0.0787467002 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000422_090 |
0.0788114465 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002734_020 |
0.0792547802 |
- |
- |
PREDICTED: uncharacterized protein LOC105631540 [Jatropha curcas] |
| 11 |
Hb_002785_020 |
0.0799314198 |
- |
- |
PREDICTED: laccase-4-like [Jatropha curcas] |
| 12 |
Hb_003788_030 |
0.0840566438 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_000462_120 |
0.085605355 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105642310 [Jatropha curcas] |
| 14 |
Hb_000594_010 |
0.0864050525 |
- |
- |
PREDICTED: lysine histidine transporter-like 6 isoform X3 [Populus euphratica] |
| 15 |
Hb_004030_040 |
0.0874585895 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 2.8 [Jatropha curcas] |
| 16 |
Hb_002757_030 |
0.0874856367 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_002636_170 |
0.0908978844 |
- |
- |
DNA-3-methyladenine glycosylase, putative [Ricinus communis] |
| 18 |
Hb_009164_010 |
0.0916256872 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_000696_130 |
0.0928030175 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 20 |
Hb_004884_040 |
0.0929541789 |
- |
- |
hypothetical protein POPTR_0016s12910g [Populus trichocarpa] |