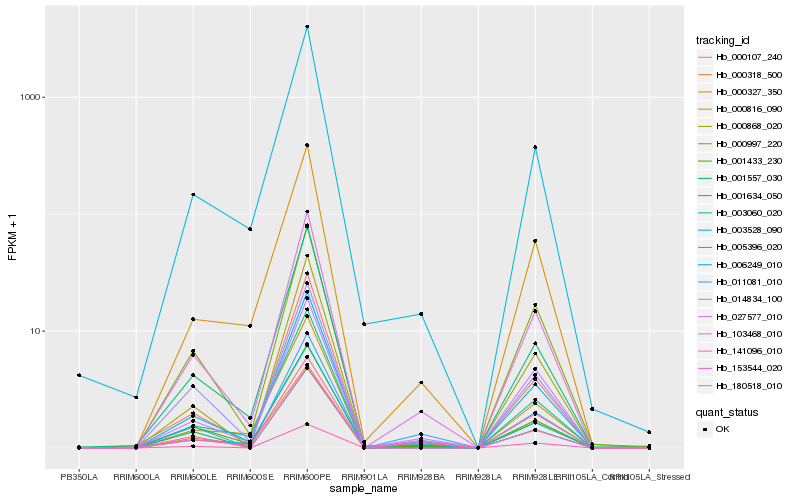
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_103468_010 |
0.0 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 2 |
Hb_003060_020 |
0.0565223647 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 3 |
Hb_014834_100 |
0.065579592 |
- |
- |
PREDICTED: probable aquaporin TIP-type [Gossypium raimondii] |
| 4 |
Hb_006249_010 |
0.0675403708 |
- |
- |
hypothetical protein JCGZ_13152 [Jatropha curcas] |
| 5 |
Hb_027577_010 |
0.0701448875 |
- |
- |
PREDICTED: uncharacterized protein LOC104779928 [Camelina sativa] |
| 6 |
Hb_000868_020 |
0.0708055072 |
- |
- |
- |
| 7 |
Hb_180518_010 |
0.0708380902 |
- |
- |
PREDICTED: receptor-like protein 2 [Jatropha curcas] |
| 8 |
Hb_003528_090 |
0.0726668798 |
- |
- |
Uncharacterized protein TCM_037169 [Theobroma cacao] |
| 9 |
Hb_000107_240 |
0.0778893672 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 10 |
Hb_000318_500 |
0.081344777 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001634_050 |
0.0814601221 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 12 [Jatropha curcas] |
| 12 |
Hb_000327_350 |
0.0822517302 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 21-like [Jatropha curcas] |
| 13 |
Hb_001433_230 |
0.0833407105 |
- |
- |
PREDICTED: kinesin-4-like [Jatropha curcas] |
| 14 |
Hb_141096_010 |
0.0841561476 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 55-like [Jatropha curcas] |
| 15 |
Hb_000816_090 |
0.0847032837 |
transcription factor |
TF Family: TRAF |
protein binding protein, putative [Ricinus communis] |
| 16 |
Hb_011081_010 |
0.0864560916 |
- |
- |
unnamed protein product [Coffea canephora] |
| 17 |
Hb_153544_020 |
0.0881012171 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP12 [Jatropha curcas] |
| 18 |
Hb_005396_020 |
0.0881433573 |
- |
- |
hypothetical protein CICLE_v10033278mg [Citrus clementina] |
| 19 |
Hb_000997_220 |
0.0899541732 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA14-like [Jatropha curcas] |
| 20 |
Hb_001557_030 |
0.090385258 |
- |
- |
hypothetical protein CICLE_v10005306mg [Citrus clementina] |