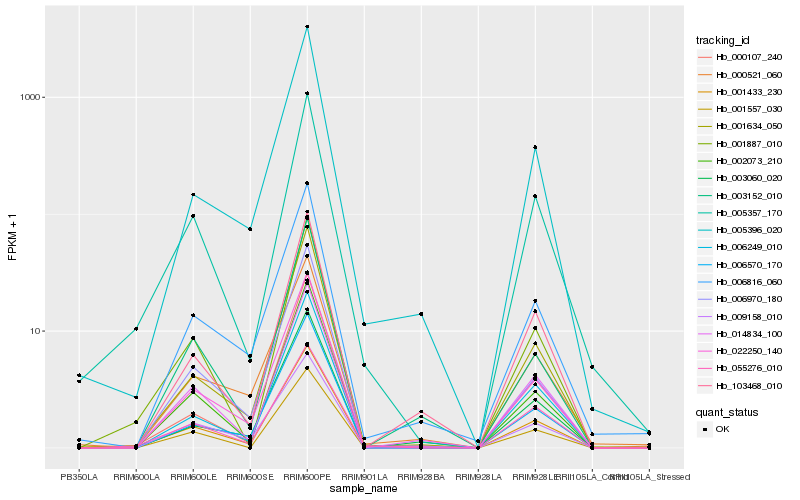
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001433_230 |
0.0 |
- |
- |
PREDICTED: kinesin-4-like [Jatropha curcas] |
| 2 |
Hb_022250_140 |
0.0642136699 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 3 |
Hb_014834_100 |
0.0686728178 |
- |
- |
PREDICTED: probable aquaporin TIP-type [Gossypium raimondii] |
| 4 |
Hb_006816_060 |
0.0693205039 |
- |
- |
PREDICTED: phospholipid-metabolizing enzyme A-C1-like [Jatropha curcas] |
| 5 |
Hb_001634_050 |
0.0791812439 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 12 [Jatropha curcas] |
| 6 |
Hb_005357_170 |
0.0809158944 |
- |
- |
Pectinesterase precursor, putative [Ricinus communis] |
| 7 |
Hb_005396_020 |
0.081108867 |
- |
- |
hypothetical protein CICLE_v10033278mg [Citrus clementina] |
| 8 |
Hb_103468_010 |
0.0833407105 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 9 |
Hb_000521_060 |
0.084370041 |
- |
- |
PREDICTED: protein YLS3-like [Jatropha curcas] |
| 10 |
Hb_002073_210 |
0.0866039762 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003060_020 |
0.0901837758 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 12 |
Hb_055276_010 |
0.0923154598 |
- |
- |
hypothetical protein POPTR_0003s02950g [Populus trichocarpa] |
| 13 |
Hb_001557_030 |
0.0928734734 |
- |
- |
hypothetical protein CICLE_v10005306mg [Citrus clementina] |
| 14 |
Hb_006970_180 |
0.0931818547 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT100-like [Jatropha curcas] |
| 15 |
Hb_006570_170 |
0.0946173839 |
- |
- |
Gibberellin-regulated protein 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_003152_010 |
0.0948029563 |
- |
- |
- |
| 17 |
Hb_006249_010 |
0.095106968 |
- |
- |
hypothetical protein JCGZ_13152 [Jatropha curcas] |
| 18 |
Hb_001887_010 |
0.0956913765 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 19 |
Hb_000107_240 |
0.0975431334 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 20 |
Hb_009158_010 |
0.0975614774 |
- |
- |
PREDICTED: cytochrome P450 78A9 [Jatropha curcas] |