| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
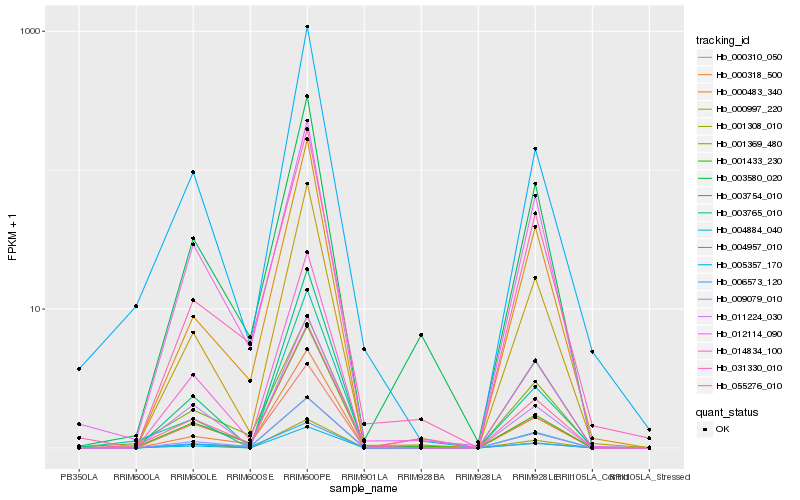
Hb_055276_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0003s02950g [Populus trichocarpa] |
| 2 |
Hb_009079_010 |
0.0550508894 |
- |
- |
JHL18I08.8 [Jatropha curcas] |
| 3 |
Hb_001369_480 |
0.0779204011 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0012s14660g [Populus trichocarpa] |
| 4 |
Hb_005357_170 |
0.0786052788 |
- |
- |
Pectinesterase precursor, putative [Ricinus communis] |
| 5 |
Hb_000997_220 |
0.0789759746 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA14-like [Jatropha curcas] |
| 6 |
Hb_000318_500 |
0.0895355045 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001433_230 |
0.0923154598 |
- |
- |
PREDICTED: kinesin-4-like [Jatropha curcas] |
| 8 |
Hb_003754_010 |
0.0924931739 |
- |
- |
hypothetical protein POPTR_0019s09950g [Populus trichocarpa] |
| 9 |
Hb_004957_010 |
0.0960487245 |
- |
- |
PREDICTED: probable aquaporin NIP-type [Prunus mume] |
| 10 |
Hb_012114_090 |
0.0970395024 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |
| 11 |
Hb_004884_040 |
0.098043098 |
- |
- |
hypothetical protein POPTR_0016s12910g [Populus trichocarpa] |
| 12 |
Hb_001308_010 |
0.0997561403 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_014834_100 |
0.1009960943 |
- |
- |
PREDICTED: probable aquaporin TIP-type [Gossypium raimondii] |
| 14 |
Hb_003580_020 |
0.101675063 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_006573_120 |
0.1021680416 |
- |
- |
PREDICTED: DUF21 domain-containing protein At5g52790 [Jatropha curcas] |
| 16 |
Hb_000483_340 |
0.1027662861 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000310_050 |
0.1038205122 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 18 |
Hb_011224_030 |
0.1044370523 |
- |
- |
PREDICTED: oligopeptide transporter 1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_031330_010 |
0.1060346589 |
- |
- |
beta-1,3-glucuronyltransferase, putative [Ricinus communis] |
| 20 |
Hb_003765_010 |
0.106714425 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |