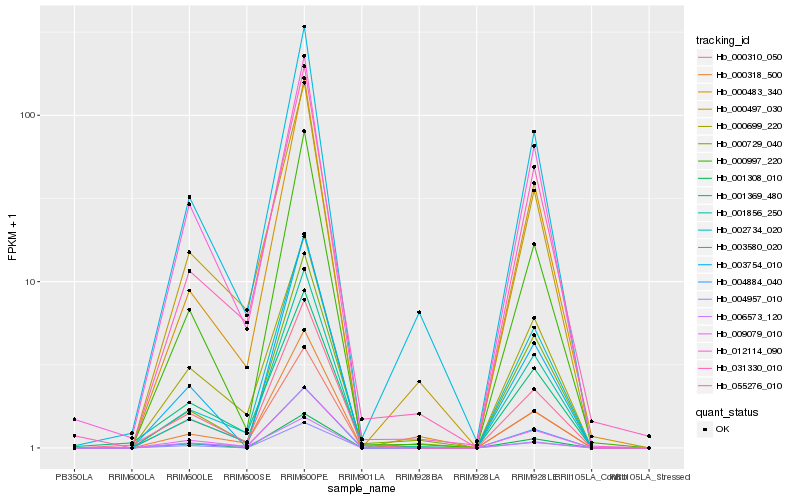
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009079_010 |
0.0 |
- |
- |
JHL18I08.8 [Jatropha curcas] |
| 2 |
Hb_055276_010 |
0.0550508894 |
- |
- |
hypothetical protein POPTR_0003s02950g [Populus trichocarpa] |
| 3 |
Hb_000997_220 |
0.072766871 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA14-like [Jatropha curcas] |
| 4 |
Hb_004884_040 |
0.0771344814 |
- |
- |
hypothetical protein POPTR_0016s12910g [Populus trichocarpa] |
| 5 |
Hb_031330_010 |
0.0804734623 |
- |
- |
beta-1,3-glucuronyltransferase, putative [Ricinus communis] |
| 6 |
Hb_012114_090 |
0.0822071695 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |
| 7 |
Hb_000318_500 |
0.0822445163 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000483_340 |
0.0845460441 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001369_480 |
0.0849342037 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0012s14660g [Populus trichocarpa] |
| 10 |
Hb_000699_220 |
0.0906931496 |
- |
- |
PREDICTED: phospholipid-metabolizing enzyme A-C1-like [Jatropha curcas] |
| 11 |
Hb_001856_250 |
0.0912480338 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Jatropha curcas] |
| 12 |
Hb_002734_020 |
0.0923733563 |
- |
- |
PREDICTED: uncharacterized protein LOC105631540 [Jatropha curcas] |
| 13 |
Hb_000497_030 |
0.0935043054 |
- |
- |
- |
| 14 |
Hb_003580_020 |
0.0939345217 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003754_010 |
0.0943939917 |
- |
- |
hypothetical protein POPTR_0019s09950g [Populus trichocarpa] |
| 16 |
Hb_000310_050 |
0.0946750707 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 17 |
Hb_001308_010 |
0.0969693293 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004957_010 |
0.0971742816 |
- |
- |
PREDICTED: probable aquaporin NIP-type [Prunus mume] |
| 19 |
Hb_006573_120 |
0.0979090557 |
- |
- |
PREDICTED: DUF21 domain-containing protein At5g52790 [Jatropha curcas] |
| 20 |
Hb_000729_040 |
0.0985119697 |
- |
- |
xyloglucan endotransglucosylase/hydrolase protein 10 precursor [Populus trichocarpa] |