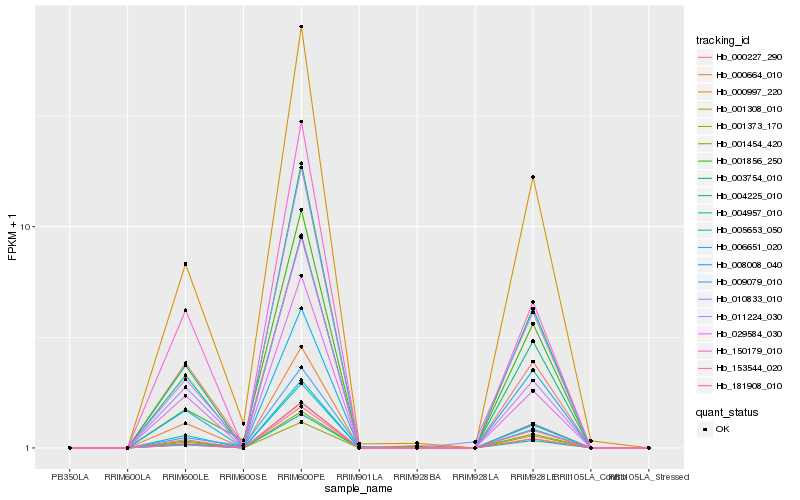
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001308_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004957_010 |
0.032949347 |
- |
- |
PREDICTED: probable aquaporin NIP-type [Prunus mume] |
| 3 |
Hb_004225_010 |
0.0397205146 |
- |
- |
- |
| 4 |
Hb_005653_050 |
0.0414106937 |
- |
- |
PREDICTED: CMP-N-acetylneuraminate-beta-1,4-galactoside alpha-2,3-sialyltransferase [Jatropha curcas] |
| 5 |
Hb_003754_010 |
0.0475513674 |
- |
- |
hypothetical protein POPTR_0019s09950g [Populus trichocarpa] |
| 6 |
Hb_000997_220 |
0.0599718546 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA14-like [Jatropha curcas] |
| 7 |
Hb_001373_170 |
0.0626573496 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 9 [Jatropha curcas] |
| 8 |
Hb_029584_030 |
0.0687561249 |
- |
- |
hypothetical protein JCGZ_07310 [Jatropha curcas] |
| 9 |
Hb_153544_020 |
0.0734808192 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP12 [Jatropha curcas] |
| 10 |
Hb_006651_020 |
0.0762396046 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 11 |
Hb_000664_010 |
0.0821935263 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 4-like [Jatropha curcas] |
| 12 |
Hb_001454_420 |
0.083224278 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 13 |
Hb_010833_010 |
0.0858214294 |
- |
- |
hypothetical protein JCGZ_05701 [Jatropha curcas] |
| 14 |
Hb_150179_010 |
0.0863577582 |
- |
- |
hypothetical protein JCGZ_21079 [Jatropha curcas] |
| 15 |
Hb_008008_040 |
0.0943877775 |
- |
- |
PREDICTED: auxin-induced protein 15A-like [Jatropha curcas] |
| 16 |
Hb_011224_030 |
0.0963534314 |
- |
- |
PREDICTED: oligopeptide transporter 1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000227_290 |
0.0964669003 |
- |
- |
PREDICTED: uncharacterized protein LOC105631957 [Jatropha curcas] |
| 18 |
Hb_009079_010 |
0.0969693293 |
- |
- |
JHL18I08.8 [Jatropha curcas] |
| 19 |
Hb_001856_250 |
0.0972089003 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Jatropha curcas] |
| 20 |
Hb_181908_010 |
0.0981911809 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73C3 [Jatropha curcas] |