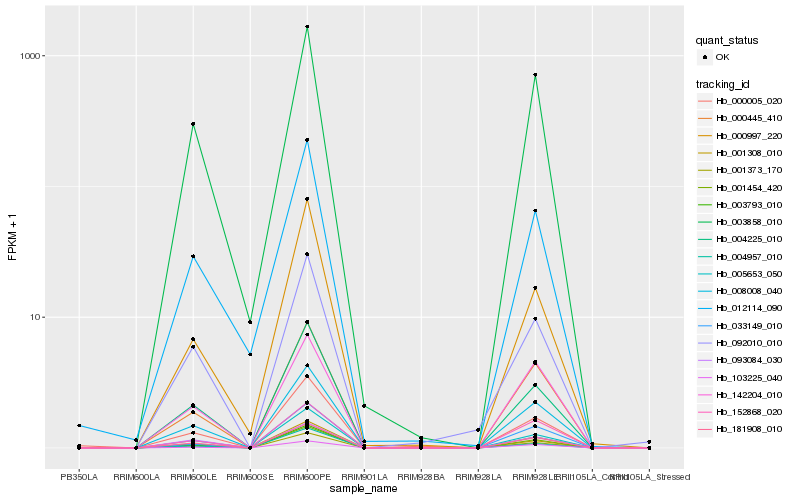
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001373_170 |
0.0 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 9 [Jatropha curcas] |
| 2 |
Hb_001454_420 |
0.0288612059 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 3 |
Hb_008008_040 |
0.031844451 |
- |
- |
PREDICTED: auxin-induced protein 15A-like [Jatropha curcas] |
| 4 |
Hb_005653_050 |
0.0356824166 |
- |
- |
PREDICTED: CMP-N-acetylneuraminate-beta-1,4-galactoside alpha-2,3-sialyltransferase [Jatropha curcas] |
| 5 |
Hb_181908_010 |
0.0387720076 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73C3 [Jatropha curcas] |
| 6 |
Hb_004225_010 |
0.0445831219 |
- |
- |
- |
| 7 |
Hb_000445_410 |
0.0572771644 |
- |
- |
hypothetical protein EUGRSUZ_F02776 [Eucalyptus grandis] |
| 8 |
Hb_003793_010 |
0.061673397 |
- |
- |
hypothetical protein JCGZ_27055 [Jatropha curcas] |
| 9 |
Hb_001308_010 |
0.0626573496 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003858_010 |
0.0742003155 |
- |
- |
xyloglucan endotransglucosylase/hydrolase [Gossypium hirsutum] |
| 11 |
Hb_103225_040 |
0.0857469783 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 12-like [Jatropha curcas] |
| 12 |
Hb_092010_010 |
0.0908204941 |
- |
- |
cysteine proteinase inhibitor 5-like [Cucumis sativus] |
| 13 |
Hb_012114_090 |
0.09143782 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |
| 14 |
Hb_152868_020 |
0.0924917337 |
- |
- |
RecName: Full=2-methylbutanal oxime monooxygenase; AltName: Full=Cytochrome P450 71E7 [Manihot esculenta] |
| 15 |
Hb_004957_010 |
0.0942488258 |
- |
- |
PREDICTED: probable aquaporin NIP-type [Prunus mume] |
| 16 |
Hb_033149_010 |
0.0946290983 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 17 |
Hb_000005_020 |
0.1011658226 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_093084_030 |
0.1024924495 |
- |
- |
oleosin2 [Plukenetia volubilis] |
| 19 |
Hb_142204_010 |
0.1041660733 |
- |
- |
pyruvate decarboxylase [Saccharum officinarum] |
| 20 |
Hb_000997_220 |
0.1043767622 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA14-like [Jatropha curcas] |