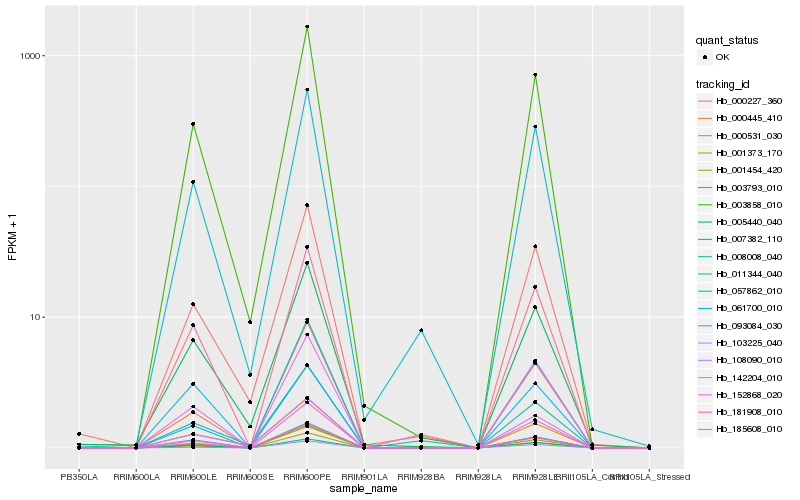
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003858_010 |
0.0 |
- |
- |
xyloglucan endotransglucosylase/hydrolase [Gossypium hirsutum] |
| 2 |
Hb_181908_010 |
0.0490443878 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73C3 [Jatropha curcas] |
| 3 |
Hb_008008_040 |
0.0521695303 |
- |
- |
PREDICTED: auxin-induced protein 15A-like [Jatropha curcas] |
| 4 |
Hb_001454_420 |
0.0595353652 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 5 |
Hb_000227_360 |
0.0673462291 |
- |
- |
PREDICTED: mavicyanin [Jatropha curcas] |
| 6 |
Hb_142204_010 |
0.0688903461 |
- |
- |
pyruvate decarboxylase [Saccharum officinarum] |
| 7 |
Hb_011344_040 |
0.0707603096 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 8 |
Hb_005440_040 |
0.0709098871 |
- |
- |
PREDICTED: microtubule-associated protein 70-5 [Jatropha curcas] |
| 9 |
Hb_185608_010 |
0.0712209564 |
- |
- |
hypothetical protein POPTR_0004s22210g [Populus trichocarpa] |
| 10 |
Hb_108090_010 |
0.0727246924 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001373_170 |
0.0742003155 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 9 [Jatropha curcas] |
| 12 |
Hb_000445_410 |
0.0747323257 |
- |
- |
hypothetical protein EUGRSUZ_F02776 [Eucalyptus grandis] |
| 13 |
Hb_003793_010 |
0.0780945642 |
- |
- |
hypothetical protein JCGZ_27055 [Jatropha curcas] |
| 14 |
Hb_093084_030 |
0.0801529143 |
- |
- |
oleosin2 [Plukenetia volubilis] |
| 15 |
Hb_152868_020 |
0.0839224292 |
- |
- |
RecName: Full=2-methylbutanal oxime monooxygenase; AltName: Full=Cytochrome P450 71E7 [Manihot esculenta] |
| 16 |
Hb_000531_030 |
0.0841538743 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_007382_110 |
0.0852277963 |
- |
- |
PREDICTED: bidirectional sugar transporter N3 [Jatropha curcas] |
| 18 |
Hb_061700_010 |
0.0861997003 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Prunus mume] |
| 19 |
Hb_057862_010 |
0.0899364213 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 20 |
Hb_103225_040 |
0.0905805571 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 12-like [Jatropha curcas] |