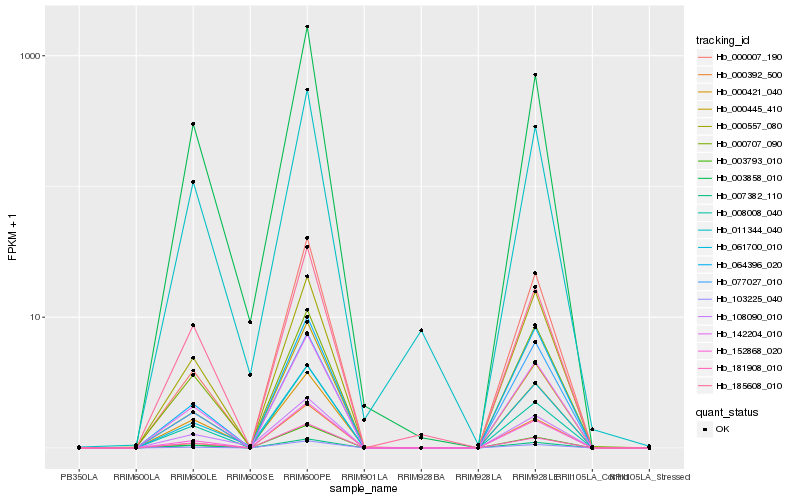
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_142204_010 |
0.0 |
- |
- |
pyruvate decarboxylase [Saccharum officinarum] |
| 2 |
Hb_152868_020 |
0.0478195493 |
- |
- |
RecName: Full=2-methylbutanal oxime monooxygenase; AltName: Full=Cytochrome P450 71E7 [Manihot esculenta] |
| 3 |
Hb_000557_080 |
0.0585629473 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_007382_110 |
0.0591984755 |
- |
- |
PREDICTED: bidirectional sugar transporter N3 [Jatropha curcas] |
| 5 |
Hb_061700_010 |
0.0650666173 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Prunus mume] |
| 6 |
Hb_103225_040 |
0.066321995 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 12-like [Jatropha curcas] |
| 7 |
Hb_003858_010 |
0.0688903461 |
- |
- |
xyloglucan endotransglucosylase/hydrolase [Gossypium hirsutum] |
| 8 |
Hb_000445_410 |
0.0692817773 |
- |
- |
hypothetical protein EUGRSUZ_F02776 [Eucalyptus grandis] |
| 9 |
Hb_003793_010 |
0.0699462085 |
- |
- |
hypothetical protein JCGZ_27055 [Jatropha curcas] |
| 10 |
Hb_008008_040 |
0.0733459489 |
- |
- |
PREDICTED: auxin-induced protein 15A-like [Jatropha curcas] |
| 11 |
Hb_185608_010 |
0.0745441154 |
- |
- |
hypothetical protein POPTR_0004s22210g [Populus trichocarpa] |
| 12 |
Hb_000707_090 |
0.0777218554 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.6-like [Jatropha curcas] |
| 13 |
Hb_181908_010 |
0.077844102 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73C3 [Jatropha curcas] |
| 14 |
Hb_011344_040 |
0.0795863899 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 15 |
Hb_064396_020 |
0.0871358686 |
- |
- |
hypothetical protein POPTR_0019s09950g [Populus trichocarpa] |
| 16 |
Hb_000421_040 |
0.087865823 |
- |
- |
Peptide transporter, putative [Ricinus communis] |
| 17 |
Hb_000007_190 |
0.0879265397 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 10-like [Jatropha curcas] |
| 18 |
Hb_108090_010 |
0.0883942999 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000392_500 |
0.0886503165 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_077027_010 |
0.091467053 |
- |
- |
hypothetical protein JCGZ_04511 [Jatropha curcas] |