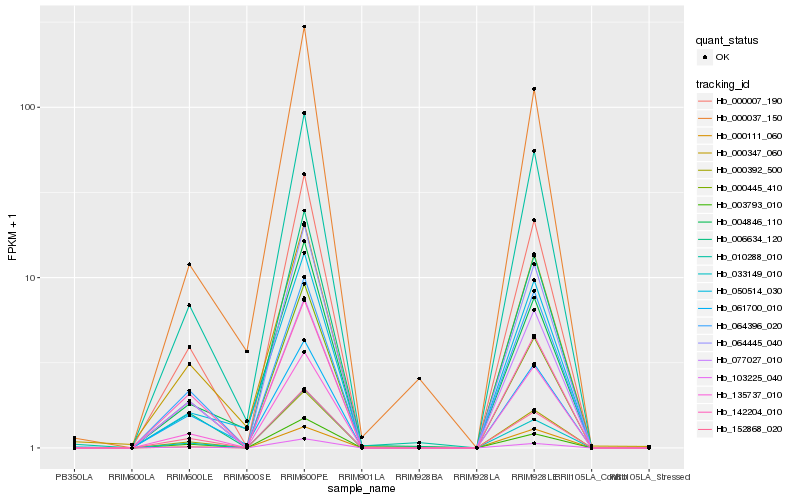
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000392_500 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000007_190 |
0.0252926474 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 10-like [Jatropha curcas] |
| 3 |
Hb_064445_040 |
0.0373036448 |
- |
- |
hypothetical protein POPTR_0005s09430g [Populus trichocarpa] |
| 4 |
Hb_010288_010 |
0.046119971 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 5 |
Hb_103225_040 |
0.049276509 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 12-like [Jatropha curcas] |
| 6 |
Hb_152868_020 |
0.0503670633 |
- |
- |
RecName: Full=2-methylbutanal oxime monooxygenase; AltName: Full=Cytochrome P450 71E7 [Manihot esculenta] |
| 7 |
Hb_135737_010 |
0.0556204696 |
- |
- |
hypothetical protein POPTR_0004s02760g [Populus trichocarpa] |
| 8 |
Hb_003793_010 |
0.0732486995 |
- |
- |
hypothetical protein JCGZ_27055 [Jatropha curcas] |
| 9 |
Hb_000445_410 |
0.0777491076 |
- |
- |
hypothetical protein EUGRSUZ_F02776 [Eucalyptus grandis] |
| 10 |
Hb_064396_020 |
0.0788288918 |
- |
- |
hypothetical protein POPTR_0019s09950g [Populus trichocarpa] |
| 11 |
Hb_006634_120 |
0.0792162375 |
- |
- |
PREDICTED: protein trichome birefringence-like 3 [Jatropha curcas] |
| 12 |
Hb_033149_010 |
0.0797281026 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 13 |
Hb_077027_010 |
0.084750812 |
- |
- |
hypothetical protein JCGZ_04511 [Jatropha curcas] |
| 14 |
Hb_142204_010 |
0.0886503165 |
- |
- |
pyruvate decarboxylase [Saccharum officinarum] |
| 15 |
Hb_000347_060 |
0.0948005635 |
- |
- |
PREDICTED: uncharacterized protein LOC105631356 [Jatropha curcas] |
| 16 |
Hb_050514_030 |
0.096457853 |
- |
- |
PREDICTED: protein trichome birefringence-like 34 [Jatropha curcas] |
| 17 |
Hb_000111_060 |
0.0981391787 |
transcription factor |
TF Family: SRS |
PREDICTED: protein SHI RELATED SEQUENCE 6 isoform X2 [Jatropha curcas] |
| 18 |
Hb_004846_110 |
0.099582363 |
- |
- |
hypothetical protein JCGZ_04591 [Jatropha curcas] |
| 19 |
Hb_000037_150 |
0.1036652383 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 20 |
Hb_061700_010 |
0.1041342235 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Prunus mume] |