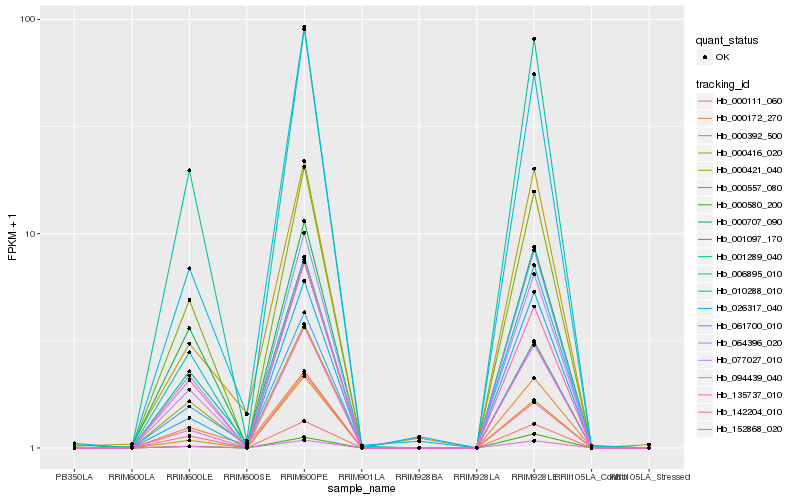
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_077027_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_04511 [Jatropha curcas] |
| 2 |
Hb_064396_020 |
0.0062088043 |
- |
- |
hypothetical protein POPTR_0019s09950g [Populus trichocarpa] |
| 3 |
Hb_094439_040 |
0.047438473 |
- |
- |
PREDICTED: cytochrome P450 93A1-like [Citrus sinensis] |
| 4 |
Hb_135737_010 |
0.0488971265 |
- |
- |
hypothetical protein POPTR_0004s02760g [Populus trichocarpa] |
| 5 |
Hb_001289_040 |
0.0539233405 |
- |
- |
NDR1/HIN1-like 1 [Theobroma cacao] |
| 6 |
Hb_000557_080 |
0.0568626619 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000392_500 |
0.084750812 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001097_170 |
0.0849866111 |
- |
- |
PREDICTED: protein NDR1 [Jatropha curcas] |
| 9 |
Hb_061700_010 |
0.0889110716 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Prunus mume] |
| 10 |
Hb_000172_270 |
0.0889737324 |
- |
- |
hypothetical protein JCGZ_24087 [Jatropha curcas] |
| 11 |
Hb_000111_060 |
0.0899286699 |
transcription factor |
TF Family: SRS |
PREDICTED: protein SHI RELATED SEQUENCE 6 isoform X2 [Jatropha curcas] |
| 12 |
Hb_142204_010 |
0.091467053 |
- |
- |
pyruvate decarboxylase [Saccharum officinarum] |
| 13 |
Hb_000421_040 |
0.0928160124 |
- |
- |
Peptide transporter, putative [Ricinus communis] |
| 14 |
Hb_006895_010 |
0.096084163 |
- |
- |
PREDICTED: secologanin synthase-like [Citrus sinensis] |
| 15 |
Hb_000416_020 |
0.0967233873 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_152868_020 |
0.0968206021 |
- |
- |
RecName: Full=2-methylbutanal oxime monooxygenase; AltName: Full=Cytochrome P450 71E7 [Manihot esculenta] |
| 17 |
Hb_000707_090 |
0.0974177408 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.6-like [Jatropha curcas] |
| 18 |
Hb_026317_040 |
0.0975312037 |
- |
- |
hypothetical protein POPTR_0009s12270g [Populus trichocarpa] |
| 19 |
Hb_010288_010 |
0.0981371521 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 20 |
Hb_000580_200 |
0.0990991774 |
- |
- |
PREDICTED: pectinesterase 2 isoform X1 [Jatropha curcas] |