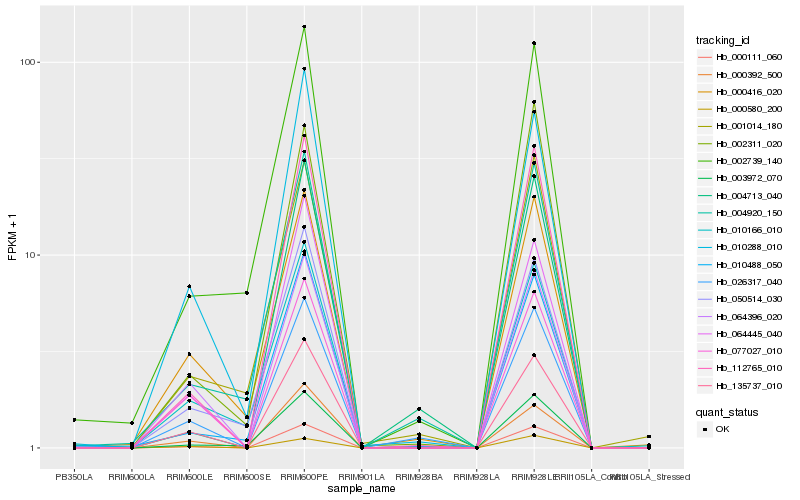
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000111_060 |
0.0 |
transcription factor |
TF Family: SRS |
PREDICTED: protein SHI RELATED SEQUENCE 6 isoform X2 [Jatropha curcas] |
| 2 |
Hb_112765_010 |
0.0417277007 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_135737_010 |
0.0537495904 |
- |
- |
hypothetical protein POPTR_0004s02760g [Populus trichocarpa] |
| 4 |
Hb_003972_070 |
0.0868196402 |
- |
- |
PREDICTED: cytochrome P450 85A1-like [Citrus sinensis] |
| 5 |
Hb_010488_050 |
0.0876443961 |
transcription factor |
TF Family: LIM |
hypothetical protein JCGZ_14428 [Jatropha curcas] |
| 6 |
Hb_026317_040 |
0.0884766632 |
- |
- |
hypothetical protein POPTR_0009s12270g [Populus trichocarpa] |
| 7 |
Hb_064396_020 |
0.0895851169 |
- |
- |
hypothetical protein POPTR_0019s09950g [Populus trichocarpa] |
| 8 |
Hb_077027_010 |
0.0899286699 |
- |
- |
hypothetical protein JCGZ_04511 [Jatropha curcas] |
| 9 |
Hb_064445_040 |
0.0906522777 |
- |
- |
hypothetical protein POPTR_0005s09430g [Populus trichocarpa] |
| 10 |
Hb_000392_500 |
0.0981391787 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_010288_010 |
0.0985312621 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 12 |
Hb_050514_030 |
0.102029375 |
- |
- |
PREDICTED: protein trichome birefringence-like 34 [Jatropha curcas] |
| 13 |
Hb_004920_150 |
0.1021374417 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002311_020 |
0.1034813161 |
- |
- |
PREDICTED: putative beta-D-xylosidase [Jatropha curcas] |
| 15 |
Hb_000416_020 |
0.1053776819 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_010166_010 |
0.1070734012 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14 [Jatropha curcas] |
| 17 |
Hb_004713_040 |
0.1082295462 |
- |
- |
hypothetical protein JCGZ_22567 [Jatropha curcas] |
| 18 |
Hb_002739_140 |
0.1091356618 |
- |
- |
PREDICTED: protein TPX2 [Jatropha curcas] |
| 19 |
Hb_001014_180 |
0.1092516412 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10 [Jatropha curcas] |
| 20 |
Hb_000580_200 |
0.1108889023 |
- |
- |
PREDICTED: pectinesterase 2 isoform X1 [Jatropha curcas] |