| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
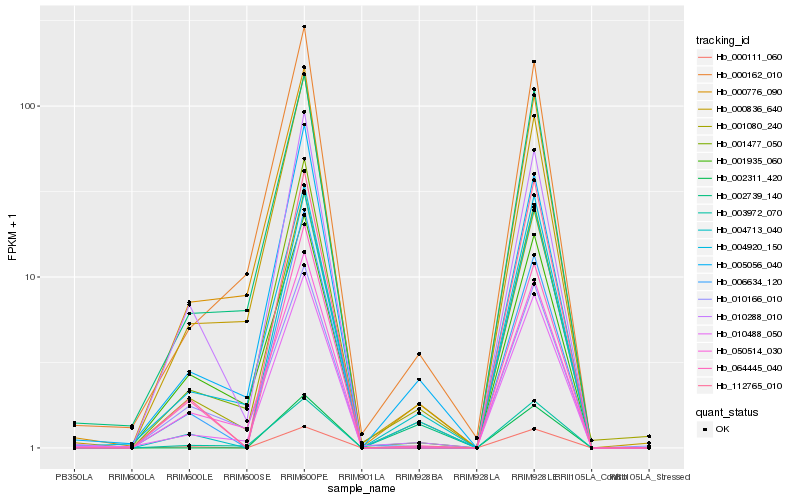
Hb_010488_050 |
0.0 |
transcription factor |
TF Family: LIM |
hypothetical protein JCGZ_14428 [Jatropha curcas] |
| 2 |
Hb_002739_140 |
0.06086913 |
- |
- |
PREDICTED: protein TPX2 [Jatropha curcas] |
| 3 |
Hb_004920_150 |
0.0672302499 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_050514_030 |
0.0708725344 |
- |
- |
PREDICTED: protein trichome birefringence-like 34 [Jatropha curcas] |
| 5 |
Hb_112765_010 |
0.075487824 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000162_010 |
0.0765943404 |
- |
- |
PREDICTED: dirigent protein 23 [Jatropha curcas] |
| 7 |
Hb_003972_070 |
0.0778634175 |
- |
- |
PREDICTED: cytochrome P450 85A1-like [Citrus sinensis] |
| 8 |
Hb_000776_090 |
0.0787428917 |
- |
- |
PREDICTED: COBRA-like protein 4 [Jatropha curcas] |
| 9 |
Hb_000836_640 |
0.0848168797 |
- |
- |
PREDICTED: uncharacterized protein LOC105644242 [Jatropha curcas] |
| 10 |
Hb_001477_050 |
0.086383778 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_000111_060 |
0.0876443961 |
transcription factor |
TF Family: SRS |
PREDICTED: protein SHI RELATED SEQUENCE 6 isoform X2 [Jatropha curcas] |
| 12 |
Hb_010166_010 |
0.0914905883 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14 [Jatropha curcas] |
| 13 |
Hb_010288_010 |
0.0932721218 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 14 |
Hb_001080_240 |
0.0951412261 |
- |
- |
hypothetical protein PHAVU_001G152800g [Phaseolus vulgaris] |
| 15 |
Hb_006634_120 |
0.0975606844 |
- |
- |
PREDICTED: protein trichome birefringence-like 3 [Jatropha curcas] |
| 16 |
Hb_004713_040 |
0.0986972698 |
- |
- |
hypothetical protein JCGZ_22567 [Jatropha curcas] |
| 17 |
Hb_064445_040 |
0.0991005348 |
- |
- |
hypothetical protein POPTR_0005s09430g [Populus trichocarpa] |
| 18 |
Hb_001935_060 |
0.0991815748 |
- |
- |
PREDICTED: metalloendoproteinase 1 [Jatropha curcas] |
| 19 |
Hb_005056_040 |
0.1013172997 |
- |
- |
hypothetical protein POPTR_0007s14730g [Populus trichocarpa] |
| 20 |
Hb_002311_420 |
0.1023630392 |
- |
- |
hypothetical protein JCGZ_18919 [Jatropha curcas] |