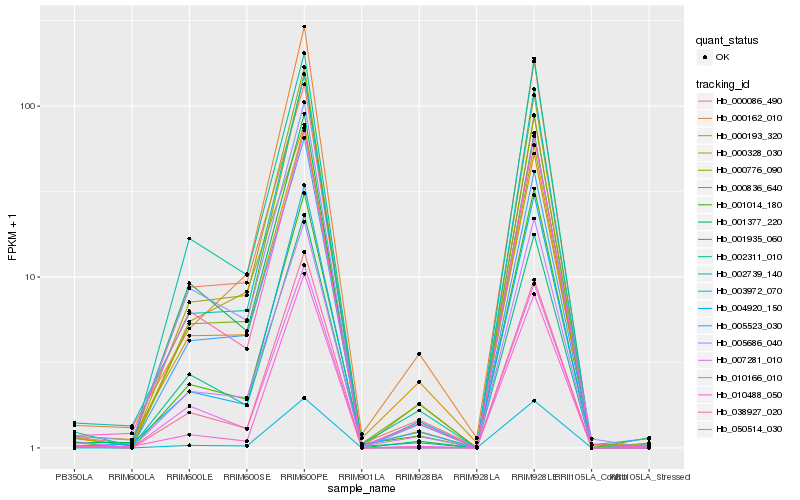
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002739_140 |
0.0 |
- |
- |
PREDICTED: protein TPX2 [Jatropha curcas] |
| 2 |
Hb_004920_150 |
0.0475412409 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000776_090 |
0.0488272382 |
- |
- |
PREDICTED: COBRA-like protein 4 [Jatropha curcas] |
| 4 |
Hb_003972_070 |
0.0533627 |
- |
- |
PREDICTED: cytochrome P450 85A1-like [Citrus sinensis] |
| 5 |
Hb_038927_020 |
0.0600681266 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC13 [Cucumis melo] |
| 6 |
Hb_010488_050 |
0.06086913 |
transcription factor |
TF Family: LIM |
hypothetical protein JCGZ_14428 [Jatropha curcas] |
| 7 |
Hb_050514_030 |
0.064723295 |
- |
- |
PREDICTED: protein trichome birefringence-like 34 [Jatropha curcas] |
| 8 |
Hb_001935_060 |
0.0655031605 |
- |
- |
PREDICTED: metalloendoproteinase 1 [Jatropha curcas] |
| 9 |
Hb_002311_010 |
0.0655192361 |
- |
- |
PREDICTED: chitinase-like protein 2 [Jatropha curcas] |
| 10 |
Hb_010166_010 |
0.0668676951 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14 [Jatropha curcas] |
| 11 |
Hb_007281_010 |
0.0693110575 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 12 |
Hb_000162_010 |
0.07648557 |
- |
- |
PREDICTED: dirigent protein 23 [Jatropha curcas] |
| 13 |
Hb_001014_180 |
0.0778035351 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10 [Jatropha curcas] |
| 14 |
Hb_001377_220 |
0.078576238 |
- |
- |
dopamine beta-monooxygenase, putative [Ricinus communis] |
| 15 |
Hb_000086_490 |
0.0794899043 |
- |
- |
cellulose synthase [Populus tomentosa] |
| 16 |
Hb_000193_320 |
0.0800420097 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005523_030 |
0.0830601394 |
- |
- |
PREDICTED: probable galacturonosyltransferase 12 [Jatropha curcas] |
| 18 |
Hb_000836_640 |
0.083480834 |
- |
- |
PREDICTED: uncharacterized protein LOC105644242 [Jatropha curcas] |
| 19 |
Hb_005686_040 |
0.086008977 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000328_030 |
0.0861205402 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 7 [UDP-forming] [Jatropha curcas] |