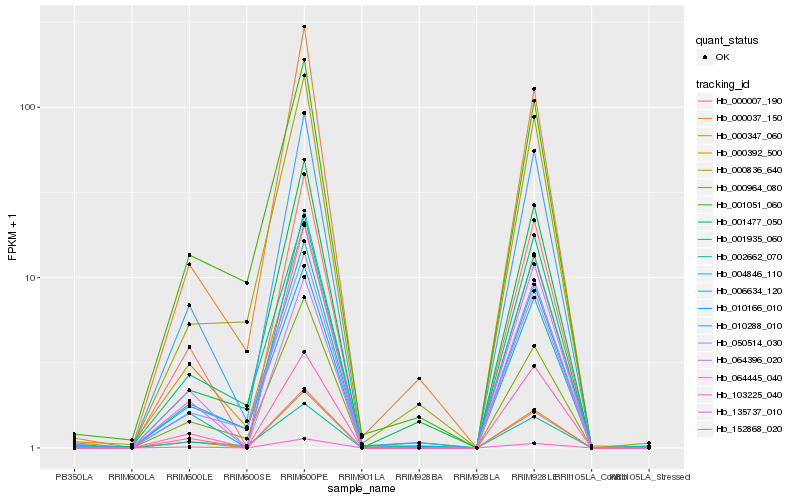
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010288_010 |
0.0 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 2 |
Hb_000392_500 |
0.046119971 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000007_190 |
0.0473639373 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 10-like [Jatropha curcas] |
| 4 |
Hb_064445_040 |
0.0512309039 |
- |
- |
hypothetical protein POPTR_0005s09430g [Populus trichocarpa] |
| 5 |
Hb_050514_030 |
0.0651863993 |
- |
- |
PREDICTED: protein trichome birefringence-like 34 [Jatropha curcas] |
| 6 |
Hb_135737_010 |
0.0677400595 |
- |
- |
hypothetical protein POPTR_0004s02760g [Populus trichocarpa] |
| 7 |
Hb_000347_060 |
0.0736135974 |
- |
- |
PREDICTED: uncharacterized protein LOC105631356 [Jatropha curcas] |
| 8 |
Hb_103225_040 |
0.0755301833 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 12-like [Jatropha curcas] |
| 9 |
Hb_006634_120 |
0.0770740749 |
- |
- |
PREDICTED: protein trichome birefringence-like 3 [Jatropha curcas] |
| 10 |
Hb_152868_020 |
0.0774645346 |
- |
- |
RecName: Full=2-methylbutanal oxime monooxygenase; AltName: Full=Cytochrome P450 71E7 [Manihot esculenta] |
| 11 |
Hb_000037_150 |
0.0813377489 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 12 |
Hb_010166_010 |
0.0814969522 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14 [Jatropha curcas] |
| 13 |
Hb_004846_110 |
0.0825140725 |
- |
- |
hypothetical protein JCGZ_04591 [Jatropha curcas] |
| 14 |
Hb_001477_050 |
0.0866391066 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_001935_060 |
0.0883328357 |
- |
- |
PREDICTED: metalloendoproteinase 1 [Jatropha curcas] |
| 16 |
Hb_000964_080 |
0.0886456485 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Jatropha curcas] |
| 17 |
Hb_002662_070 |
0.0903664086 |
- |
- |
hypothetical protein JCGZ_16102 [Jatropha curcas] |
| 18 |
Hb_001051_060 |
0.0918135571 |
- |
- |
UDP-glucuronic acid/UDP-N-acetylgalactosamine transporter, putative [Ricinus communis] |
| 19 |
Hb_000836_640 |
0.0918313951 |
- |
- |
PREDICTED: uncharacterized protein LOC105644242 [Jatropha curcas] |
| 20 |
Hb_064396_020 |
0.0932073693 |
- |
- |
hypothetical protein POPTR_0019s09950g [Populus trichocarpa] |