| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
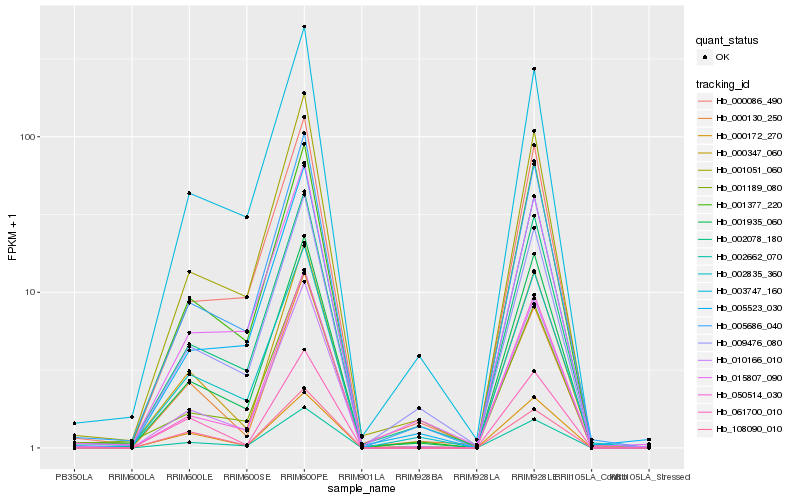
Hb_002662_070 |
0.0 |
- |
- |
hypothetical protein JCGZ_16102 [Jatropha curcas] |
| 2 |
Hb_001377_220 |
0.0500474445 |
- |
- |
dopamine beta-monooxygenase, putative [Ricinus communis] |
| 3 |
Hb_005686_040 |
0.057687171 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001051_060 |
0.0602554955 |
- |
- |
UDP-glucuronic acid/UDP-N-acetylgalactosamine transporter, putative [Ricinus communis] |
| 5 |
Hb_001935_060 |
0.0638014656 |
- |
- |
PREDICTED: metalloendoproteinase 1 [Jatropha curcas] |
| 6 |
Hb_000347_060 |
0.0688819211 |
- |
- |
PREDICTED: uncharacterized protein LOC105631356 [Jatropha curcas] |
| 7 |
Hb_002835_360 |
0.0700733731 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s09430g [Populus trichocarpa] |
| 8 |
Hb_003747_160 |
0.071022946 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002078_180 |
0.0716601152 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 8 [Jatropha curcas] |
| 10 |
Hb_010166_010 |
0.0716882904 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14 [Jatropha curcas] |
| 11 |
Hb_050514_030 |
0.0746442174 |
- |
- |
PREDICTED: protein trichome birefringence-like 34 [Jatropha curcas] |
| 12 |
Hb_000086_490 |
0.0749385491 |
- |
- |
cellulose synthase [Populus tomentosa] |
| 13 |
Hb_061700_010 |
0.0759160186 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Prunus mume] |
| 14 |
Hb_005523_030 |
0.0816948315 |
- |
- |
PREDICTED: probable galacturonosyltransferase 12 [Jatropha curcas] |
| 15 |
Hb_015807_090 |
0.0833437323 |
- |
- |
PREDICTED: probable galacturonosyltransferase 12 [Jatropha curcas] |
| 16 |
Hb_000130_250 |
0.0849718815 |
- |
- |
PREDICTED: protein trichome birefringence-like 31 [Jatropha curcas] |
| 17 |
Hb_009476_080 |
0.0859111947 |
- |
- |
BnaA03g18920D [Brassica napus] |
| 18 |
Hb_108090_010 |
0.0861773124 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000172_270 |
0.0875408125 |
- |
- |
hypothetical protein JCGZ_24087 [Jatropha curcas] |
| 20 |
Hb_001189_080 |
0.088691049 |
- |
- |
conserved hypothetical protein [Ricinus communis] |