| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
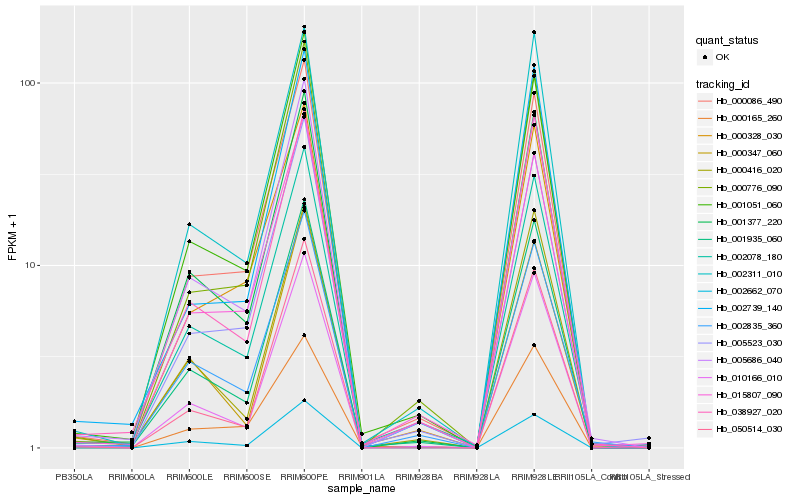
Hb_001377_220 |
0.0 |
- |
- |
dopamine beta-monooxygenase, putative [Ricinus communis] |
| 2 |
Hb_001935_060 |
0.0296516711 |
- |
- |
PREDICTED: metalloendoproteinase 1 [Jatropha curcas] |
| 3 |
Hb_010166_010 |
0.0469992923 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14 [Jatropha curcas] |
| 4 |
Hb_002311_010 |
0.0489906717 |
- |
- |
PREDICTED: chitinase-like protein 2 [Jatropha curcas] |
| 5 |
Hb_002662_070 |
0.0500474445 |
- |
- |
hypothetical protein JCGZ_16102 [Jatropha curcas] |
| 6 |
Hb_005686_040 |
0.0512986034 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002078_180 |
0.0553333555 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 8 [Jatropha curcas] |
| 8 |
Hb_038927_020 |
0.0579670388 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC13 [Cucumis melo] |
| 9 |
Hb_000086_490 |
0.0612155575 |
- |
- |
cellulose synthase [Populus tomentosa] |
| 10 |
Hb_000416_020 |
0.0637937705 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002835_360 |
0.0661615225 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s09430g [Populus trichocarpa] |
| 12 |
Hb_001051_060 |
0.0669807177 |
- |
- |
UDP-glucuronic acid/UDP-N-acetylgalactosamine transporter, putative [Ricinus communis] |
| 13 |
Hb_050514_030 |
0.0678777758 |
- |
- |
PREDICTED: protein trichome birefringence-like 34 [Jatropha curcas] |
| 14 |
Hb_005523_030 |
0.0735191033 |
- |
- |
PREDICTED: probable galacturonosyltransferase 12 [Jatropha curcas] |
| 15 |
Hb_000347_060 |
0.0739725481 |
- |
- |
PREDICTED: uncharacterized protein LOC105631356 [Jatropha curcas] |
| 16 |
Hb_000776_090 |
0.0741049321 |
- |
- |
PREDICTED: COBRA-like protein 4 [Jatropha curcas] |
| 17 |
Hb_000328_030 |
0.0744520997 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 7 [UDP-forming] [Jatropha curcas] |
| 18 |
Hb_002739_140 |
0.078576238 |
- |
- |
PREDICTED: protein TPX2 [Jatropha curcas] |
| 19 |
Hb_015807_090 |
0.0796572859 |
- |
- |
PREDICTED: probable galacturonosyltransferase 12 [Jatropha curcas] |
| 20 |
Hb_000165_260 |
0.0799266187 |
- |
- |
PREDICTED: uncharacterized protein LOC105133752 isoform X2 [Populus euphratica] |