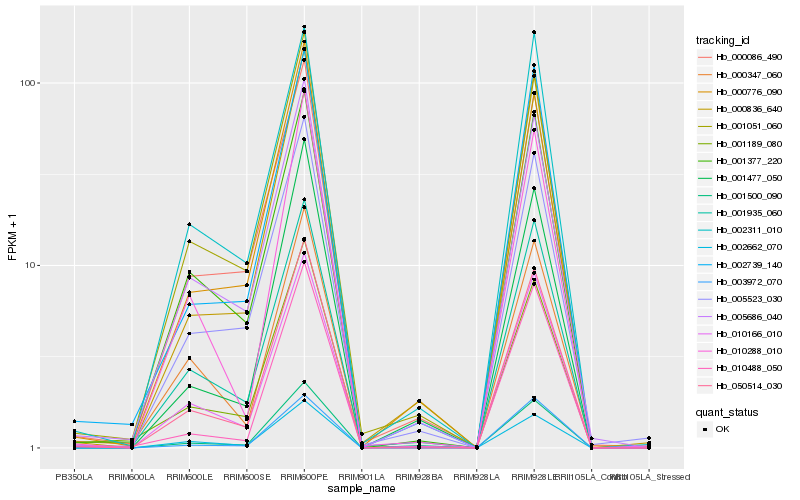
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_050514_030 |
0.0 |
- |
- |
PREDICTED: protein trichome birefringence-like 34 [Jatropha curcas] |
| 2 |
Hb_010166_010 |
0.0520067009 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14 [Jatropha curcas] |
| 3 |
Hb_000776_090 |
0.0523738756 |
- |
- |
PREDICTED: COBRA-like protein 4 [Jatropha curcas] |
| 4 |
Hb_001935_060 |
0.0595994389 |
- |
- |
PREDICTED: metalloendoproteinase 1 [Jatropha curcas] |
| 5 |
Hb_000836_640 |
0.0630106673 |
- |
- |
PREDICTED: uncharacterized protein LOC105644242 [Jatropha curcas] |
| 6 |
Hb_002739_140 |
0.064723295 |
- |
- |
PREDICTED: protein TPX2 [Jatropha curcas] |
| 7 |
Hb_010288_010 |
0.0651863993 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 8 |
Hb_005686_040 |
0.0665000093 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001377_220 |
0.0678777758 |
- |
- |
dopamine beta-monooxygenase, putative [Ricinus communis] |
| 10 |
Hb_001051_060 |
0.0685217012 |
- |
- |
UDP-glucuronic acid/UDP-N-acetylgalactosamine transporter, putative [Ricinus communis] |
| 11 |
Hb_010488_050 |
0.0708725344 |
transcription factor |
TF Family: LIM |
hypothetical protein JCGZ_14428 [Jatropha curcas] |
| 12 |
Hb_005523_030 |
0.0720083881 |
- |
- |
PREDICTED: probable galacturonosyltransferase 12 [Jatropha curcas] |
| 13 |
Hb_000086_490 |
0.0731952165 |
- |
- |
cellulose synthase [Populus tomentosa] |
| 14 |
Hb_001189_080 |
0.0738572672 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002662_070 |
0.0746442174 |
- |
- |
hypothetical protein JCGZ_16102 [Jatropha curcas] |
| 16 |
Hb_003972_070 |
0.0751862933 |
- |
- |
PREDICTED: cytochrome P450 85A1-like [Citrus sinensis] |
| 17 |
Hb_001477_050 |
0.0816590855 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 18 |
Hb_001500_090 |
0.0843133152 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002311_010 |
0.08484318 |
- |
- |
PREDICTED: chitinase-like protein 2 [Jatropha curcas] |
| 20 |
Hb_000347_060 |
0.0854443291 |
- |
- |
PREDICTED: uncharacterized protein LOC105631356 [Jatropha curcas] |