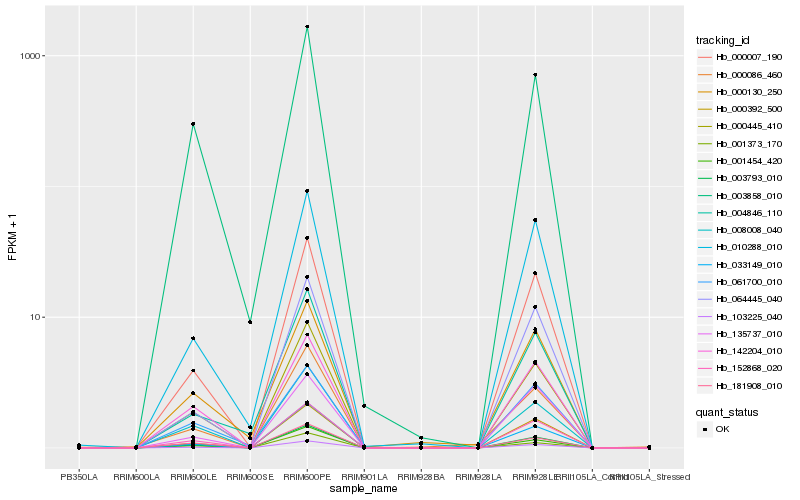
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_103225_040 |
0.0 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 12-like [Jatropha curcas] |
| 2 |
Hb_152868_020 |
0.0199650736 |
- |
- |
RecName: Full=2-methylbutanal oxime monooxygenase; AltName: Full=Cytochrome P450 71E7 [Manihot esculenta] |
| 3 |
Hb_003793_010 |
0.0241867716 |
- |
- |
hypothetical protein JCGZ_27055 [Jatropha curcas] |
| 4 |
Hb_000445_410 |
0.028578305 |
- |
- |
hypothetical protein EUGRSUZ_F02776 [Eucalyptus grandis] |
| 5 |
Hb_000007_190 |
0.0370851299 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 10-like [Jatropha curcas] |
| 6 |
Hb_000392_500 |
0.049276509 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_033149_010 |
0.0587984529 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 8 |
Hb_142204_010 |
0.066321995 |
- |
- |
pyruvate decarboxylase [Saccharum officinarum] |
| 9 |
Hb_008008_040 |
0.0701784342 |
- |
- |
PREDICTED: auxin-induced protein 15A-like [Jatropha curcas] |
| 10 |
Hb_010288_010 |
0.0755301833 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 11 |
Hb_064445_040 |
0.0759382761 |
- |
- |
hypothetical protein POPTR_0005s09430g [Populus trichocarpa] |
| 12 |
Hb_001373_170 |
0.0857469783 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 9 [Jatropha curcas] |
| 13 |
Hb_181908_010 |
0.0864016348 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73C3 [Jatropha curcas] |
| 14 |
Hb_003858_010 |
0.0905805571 |
- |
- |
xyloglucan endotransglucosylase/hydrolase [Gossypium hirsutum] |
| 15 |
Hb_004846_110 |
0.0950838884 |
- |
- |
hypothetical protein JCGZ_04591 [Jatropha curcas] |
| 16 |
Hb_001454_420 |
0.0957402585 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 17 |
Hb_061700_010 |
0.0986710419 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Prunus mume] |
| 18 |
Hb_135737_010 |
0.0997166075 |
- |
- |
hypothetical protein POPTR_0004s02760g [Populus trichocarpa] |
| 19 |
Hb_000130_250 |
0.1023824111 |
- |
- |
PREDICTED: protein trichome birefringence-like 31 [Jatropha curcas] |
| 20 |
Hb_000086_460 |
0.1044292012 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.2-like [Jatropha curcas] |