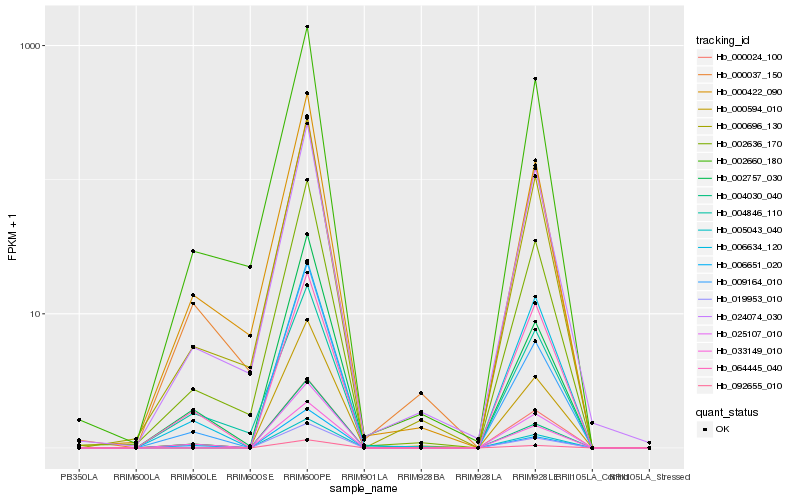
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_019953_010 |
0.0 |
- |
- |
hypothetical protein CICLE_v10019877mg [Citrus clementina] |
| 2 |
Hb_033149_010 |
0.057592479 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 3 |
Hb_002636_170 |
0.0629902446 |
- |
- |
DNA-3-methyladenine glycosylase, putative [Ricinus communis] |
| 4 |
Hb_000594_010 |
0.0669476307 |
- |
- |
PREDICTED: lysine histidine transporter-like 6 isoform X3 [Populus euphratica] |
| 5 |
Hb_000696_130 |
0.0713508369 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 6 |
Hb_006634_120 |
0.0791577155 |
- |
- |
PREDICTED: protein trichome birefringence-like 3 [Jatropha curcas] |
| 7 |
Hb_000422_090 |
0.0794940665 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_009164_010 |
0.0803608223 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_004030_040 |
0.0808922552 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 2.8 [Jatropha curcas] |
| 10 |
Hb_002660_180 |
0.0809847618 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000037_150 |
0.0830548165 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 12 |
Hb_024074_030 |
0.0928683482 |
- |
- |
hypothetical protein CISIN_1g043166mg [Citrus sinensis] |
| 13 |
Hb_006651_020 |
0.0933531794 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 14 |
Hb_025107_010 |
0.0942402534 |
- |
- |
PREDICTED: alkaline/neutral invertase CINV2-like [Sesamum indicum] |
| 15 |
Hb_005043_040 |
0.0956457555 |
- |
- |
hypothetical protein POPTR_0004s17180g [Populus trichocarpa] |
| 16 |
Hb_000024_100 |
0.0956760752 |
- |
- |
PREDICTED: uncharacterized protein LOC105631348 [Jatropha curcas] |
| 17 |
Hb_064445_040 |
0.0957136296 |
- |
- |
hypothetical protein POPTR_0005s09430g [Populus trichocarpa] |
| 18 |
Hb_004846_110 |
0.0992244835 |
- |
- |
hypothetical protein JCGZ_04591 [Jatropha curcas] |
| 19 |
Hb_092655_010 |
0.0992385714 |
- |
- |
PREDICTED: uncharacterized protein LOC105650151 [Jatropha curcas] |
| 20 |
Hb_002757_030 |
0.1014651406 |
- |
- |
unnamed protein product [Vitis vinifera] |