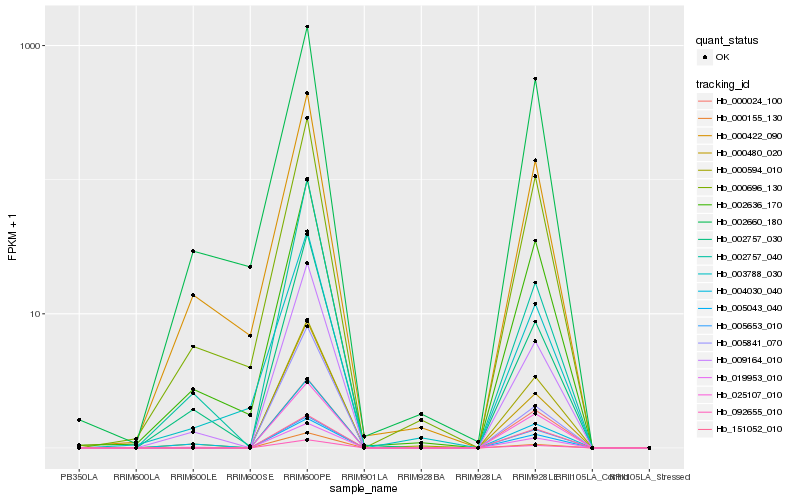
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_092655_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650151 [Jatropha curcas] |
| 2 |
Hb_025107_010 |
0.0483343348 |
- |
- |
PREDICTED: alkaline/neutral invertase CINV2-like [Sesamum indicum] |
| 3 |
Hb_005043_040 |
0.0558639209 |
- |
- |
hypothetical protein POPTR_0004s17180g [Populus trichocarpa] |
| 4 |
Hb_000024_100 |
0.055998615 |
- |
- |
PREDICTED: uncharacterized protein LOC105631348 [Jatropha curcas] |
| 5 |
Hb_000155_130 |
0.0592648856 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000480_020 |
0.0663314779 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000594_010 |
0.0701773055 |
- |
- |
PREDICTED: lysine histidine transporter-like 6 isoform X3 [Populus euphratica] |
| 8 |
Hb_009164_010 |
0.0867314173 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_019953_010 |
0.0992385714 |
- |
- |
hypothetical protein CICLE_v10019877mg [Citrus clementina] |
| 10 |
Hb_151052_010 |
0.1043387908 |
- |
- |
- |
| 11 |
Hb_005653_010 |
0.1044278059 |
- |
- |
CEN-like protein 2 [Jatropha curcas] |
| 12 |
Hb_002636_170 |
0.1045729635 |
- |
- |
DNA-3-methyladenine glycosylase, putative [Ricinus communis] |
| 13 |
Hb_005841_070 |
0.1091038301 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000696_130 |
0.1099210748 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 15 |
Hb_004030_040 |
0.1196313647 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 2.8 [Jatropha curcas] |
| 16 |
Hb_002757_030 |
0.1219019158 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_003788_030 |
0.1263385376 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 18 |
Hb_002757_040 |
0.128241869 |
- |
- |
- |
| 19 |
Hb_002660_180 |
0.1285557534 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000422_090 |
0.12860849 |
- |
- |
conserved hypothetical protein [Ricinus communis] |