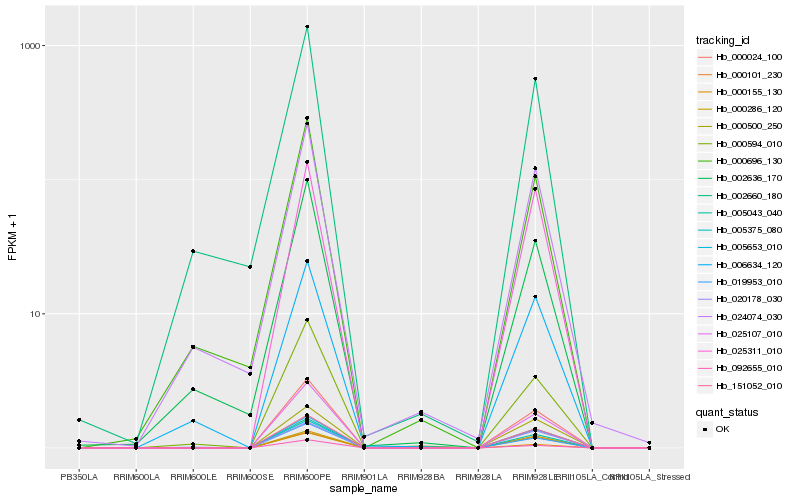
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000024_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631348 [Jatropha curcas] |
| 2 |
Hb_005043_040 |
0.0001349222 |
- |
- |
hypothetical protein POPTR_0004s17180g [Populus trichocarpa] |
| 3 |
Hb_025107_010 |
0.0076756244 |
- |
- |
PREDICTED: alkaline/neutral invertase CINV2-like [Sesamum indicum] |
| 4 |
Hb_151052_010 |
0.0484837439 |
- |
- |
- |
| 5 |
Hb_005653_010 |
0.04857313 |
- |
- |
CEN-like protein 2 [Jatropha curcas] |
| 6 |
Hb_092655_010 |
0.055998615 |
- |
- |
PREDICTED: uncharacterized protein LOC105650151 [Jatropha curcas] |
| 7 |
Hb_000500_250 |
0.0849358719 |
- |
- |
hypothetical protein CISIN_1g006096mg [Citrus sinensis] |
| 8 |
Hb_020178_030 |
0.0868779256 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000594_010 |
0.0887948427 |
- |
- |
PREDICTED: lysine histidine transporter-like 6 isoform X3 [Populus euphratica] |
| 10 |
Hb_025311_010 |
0.0890488603 |
- |
- |
- |
| 11 |
Hb_005375_080 |
0.0915895773 |
- |
- |
PREDICTED: naringenin,2-oxoglutarate 3-dioxygenase-like [Jatropha curcas] |
| 12 |
Hb_019953_010 |
0.0956760752 |
- |
- |
hypothetical protein CICLE_v10019877mg [Citrus clementina] |
| 13 |
Hb_000101_230 |
0.0989133038 |
- |
- |
hypothetical protein CICLE_v10030840mg [Citrus clementina] |
| 14 |
Hb_000286_120 |
0.0997847469 |
- |
- |
PREDICTED: uncharacterized protein LOC105633393 [Jatropha curcas] |
| 15 |
Hb_002636_170 |
0.1044390572 |
- |
- |
DNA-3-methyladenine glycosylase, putative [Ricinus communis] |
| 16 |
Hb_000696_130 |
0.1046456028 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 17 |
Hb_006634_120 |
0.1082182943 |
- |
- |
PREDICTED: protein trichome birefringence-like 3 [Jatropha curcas] |
| 18 |
Hb_024074_030 |
0.1124349349 |
- |
- |
hypothetical protein CISIN_1g043166mg [Citrus sinensis] |
| 19 |
Hb_002660_180 |
0.11399128 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000155_130 |
0.115019344 |
- |
- |
conserved hypothetical protein [Ricinus communis] |