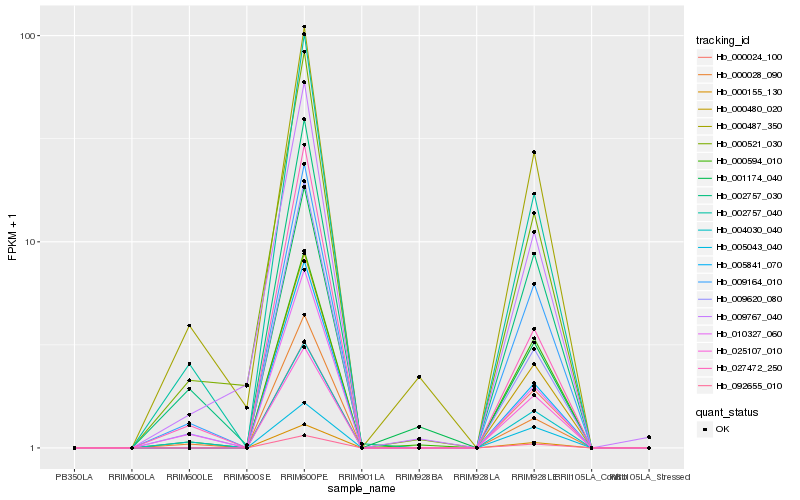
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000480_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000155_130 |
0.0070885177 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_005841_070 |
0.0430410138 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_092655_010 |
0.0663314779 |
- |
- |
PREDICTED: uncharacterized protein LOC105650151 [Jatropha curcas] |
| 5 |
Hb_009164_010 |
0.0783195228 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_002757_040 |
0.0878274607 |
- |
- |
- |
| 7 |
Hb_000594_010 |
0.0974351833 |
- |
- |
PREDICTED: lysine histidine transporter-like 6 isoform X3 [Populus euphratica] |
| 8 |
Hb_001174_040 |
0.10379551 |
- |
- |
- |
| 9 |
Hb_002757_030 |
0.1041524601 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_009620_080 |
0.1076922666 |
- |
- |
hypothetical protein POPTR_0007s08150g [Populus trichocarpa] |
| 11 |
Hb_000028_090 |
0.1092978262 |
- |
- |
PREDICTED: uncharacterized protein LOC105135810 [Populus euphratica] |
| 12 |
Hb_010327_060 |
0.1119048665 |
- |
- |
hypothetical protein B456_001G246300 [Gossypium raimondii] |
| 13 |
Hb_009767_040 |
0.1128049398 |
- |
- |
PREDICTED: uncharacterized protein LOC105127022 [Populus euphratica] |
| 14 |
Hb_004030_040 |
0.1131857857 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 2.8 [Jatropha curcas] |
| 15 |
Hb_000521_030 |
0.1138954728 |
- |
- |
PREDICTED: uncharacterized protein LOC105650350 [Jatropha curcas] |
| 16 |
Hb_025107_010 |
0.1144256212 |
- |
- |
PREDICTED: alkaline/neutral invertase CINV2-like [Sesamum indicum] |
| 17 |
Hb_027472_250 |
0.1197136385 |
- |
- |
hypothetical protein POPTR_0007s08150g [Populus trichocarpa] |
| 18 |
Hb_005043_040 |
0.1219032052 |
- |
- |
hypothetical protein POPTR_0004s17180g [Populus trichocarpa] |
| 19 |
Hb_000024_100 |
0.1220369366 |
- |
- |
PREDICTED: uncharacterized protein LOC105631348 [Jatropha curcas] |
| 20 |
Hb_000487_350 |
0.1334790311 |
- |
- |
conserved hypothetical protein [Ricinus communis] |