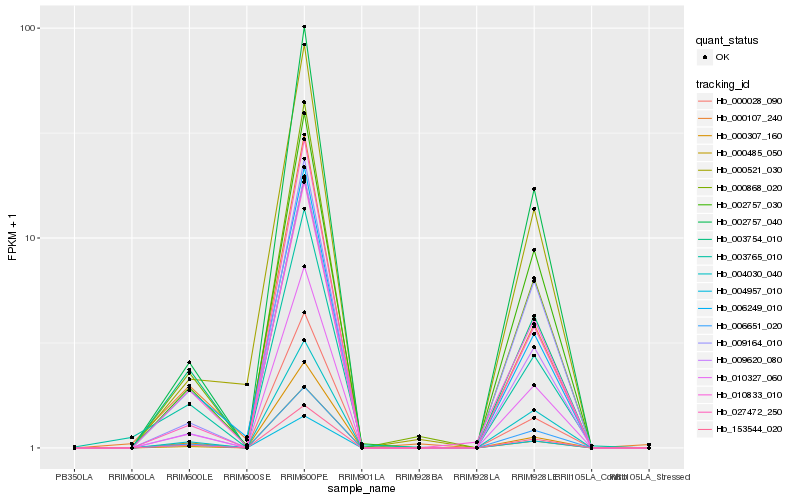
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010327_060 |
0.0 |
- |
- |
hypothetical protein B456_001G246300 [Gossypium raimondii] |
| 2 |
Hb_002757_040 |
0.029437621 |
- |
- |
- |
| 3 |
Hb_000868_020 |
0.0486865984 |
- |
- |
- |
| 4 |
Hb_002757_030 |
0.0491204711 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_153544_020 |
0.0507420923 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP12 [Jatropha curcas] |
| 6 |
Hb_000028_090 |
0.0552994529 |
- |
- |
PREDICTED: uncharacterized protein LOC105135810 [Populus euphratica] |
| 7 |
Hb_004030_040 |
0.0583713308 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 2.8 [Jatropha curcas] |
| 8 |
Hb_006651_020 |
0.0653756622 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 9 |
Hb_010833_010 |
0.0667949899 |
- |
- |
hypothetical protein JCGZ_05701 [Jatropha curcas] |
| 10 |
Hb_009164_010 |
0.0695696138 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_009620_080 |
0.0697558308 |
- |
- |
hypothetical protein POPTR_0007s08150g [Populus trichocarpa] |
| 12 |
Hb_006249_010 |
0.0717542844 |
- |
- |
hypothetical protein JCGZ_13152 [Jatropha curcas] |
| 13 |
Hb_027472_250 |
0.0771411267 |
- |
- |
hypothetical protein POPTR_0007s08150g [Populus trichocarpa] |
| 14 |
Hb_000521_030 |
0.0829410945 |
- |
- |
PREDICTED: uncharacterized protein LOC105650350 [Jatropha curcas] |
| 15 |
Hb_003754_010 |
0.0850711756 |
- |
- |
hypothetical protein POPTR_0019s09950g [Populus trichocarpa] |
| 16 |
Hb_000485_050 |
0.0861008795 |
- |
- |
PREDICTED: alpha-dioxygenase 1 [Jatropha curcas] |
| 17 |
Hb_000107_240 |
0.0876614098 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 18 |
Hb_000307_160 |
0.0879480579 |
- |
- |
PREDICTED: uncharacterized protein At1g76070 [Jatropha curcas] |
| 19 |
Hb_004957_010 |
0.0933866188 |
- |
- |
PREDICTED: probable aquaporin NIP-type [Prunus mume] |
| 20 |
Hb_003765_010 |
0.0941767253 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |