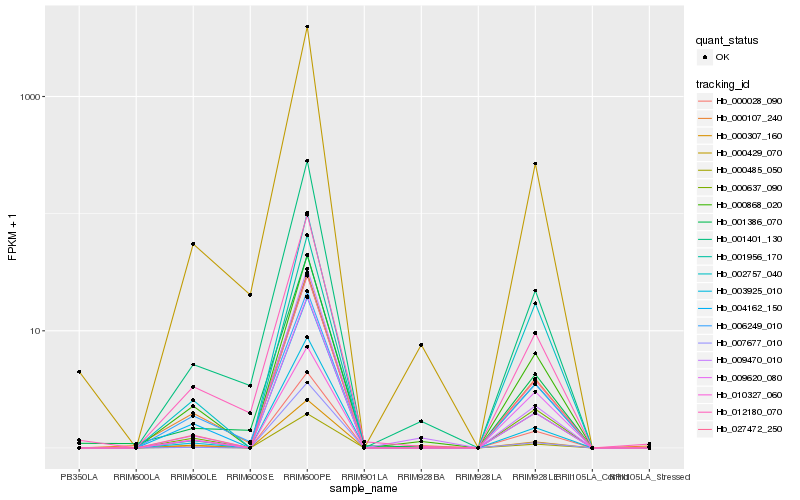
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000485_050 |
0.0 |
- |
- |
PREDICTED: alpha-dioxygenase 1 [Jatropha curcas] |
| 2 |
Hb_027472_250 |
0.0303322651 |
- |
- |
hypothetical protein POPTR_0007s08150g [Populus trichocarpa] |
| 3 |
Hb_003925_010 |
0.0416860035 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 4 |
Hb_009620_080 |
0.0422288683 |
- |
- |
hypothetical protein POPTR_0007s08150g [Populus trichocarpa] |
| 5 |
Hb_000028_090 |
0.0427522124 |
- |
- |
PREDICTED: uncharacterized protein LOC105135810 [Populus euphratica] |
| 6 |
Hb_000307_160 |
0.0498819555 |
- |
- |
PREDICTED: uncharacterized protein At1g76070 [Jatropha curcas] |
| 7 |
Hb_004162_150 |
0.061798411 |
- |
- |
hypothetical protein POPTR_0009s00870g [Populus trichocarpa] |
| 8 |
Hb_000429_070 |
0.0621825491 |
- |
- |
hypothetical protein POPTR_0006s05480g [Populus trichocarpa] |
| 9 |
Hb_001401_130 |
0.0713837256 |
- |
- |
Calcium-binding EF-hand family protein, putative [Theobroma cacao] |
| 10 |
Hb_000868_020 |
0.0720550163 |
- |
- |
- |
| 11 |
Hb_000107_240 |
0.0735176124 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 12 |
Hb_000637_090 |
0.0759153469 |
- |
- |
hypothetical protein JCGZ_05877 [Jatropha curcas] |
| 13 |
Hb_007677_010 |
0.0799441163 |
- |
- |
PREDICTED: uncharacterized protein LOC105641028 [Jatropha curcas] |
| 14 |
Hb_012180_070 |
0.0834361597 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001386_070 |
0.0858485681 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1-like [Jatropha curcas] |
| 16 |
Hb_010327_060 |
0.0861008795 |
- |
- |
hypothetical protein B456_001G246300 [Gossypium raimondii] |
| 17 |
Hb_002757_040 |
0.0865309383 |
- |
- |
- |
| 18 |
Hb_006249_010 |
0.0915415935 |
- |
- |
hypothetical protein JCGZ_13152 [Jatropha curcas] |
| 19 |
Hb_001956_170 |
0.0920957992 |
- |
- |
hypothetical protein POPTR_0001s24950g [Populus trichocarpa] |
| 20 |
Hb_009470_010 |
0.0930935084 |
- |
- |
- |