| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
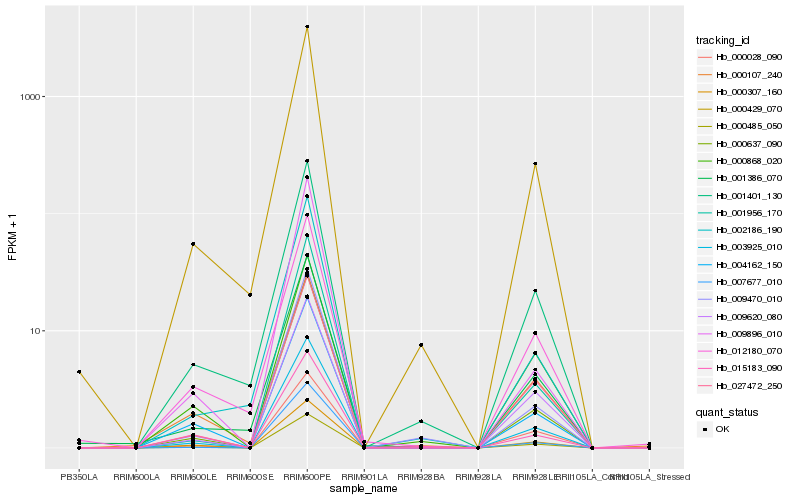
Hb_003925_010 |
0.0 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 2 |
Hb_000485_050 |
0.0416860035 |
- |
- |
PREDICTED: alpha-dioxygenase 1 [Jatropha curcas] |
| 3 |
Hb_000637_090 |
0.0553971244 |
- |
- |
hypothetical protein JCGZ_05877 [Jatropha curcas] |
| 4 |
Hb_027472_250 |
0.0582705365 |
- |
- |
hypothetical protein POPTR_0007s08150g [Populus trichocarpa] |
| 5 |
Hb_007677_010 |
0.060678935 |
- |
- |
PREDICTED: uncharacterized protein LOC105641028 [Jatropha curcas] |
| 6 |
Hb_004162_150 |
0.0646270086 |
- |
- |
hypothetical protein POPTR_0009s00870g [Populus trichocarpa] |
| 7 |
Hb_000429_070 |
0.0659757175 |
- |
- |
hypothetical protein POPTR_0006s05480g [Populus trichocarpa] |
| 8 |
Hb_009620_080 |
0.0699554304 |
- |
- |
hypothetical protein POPTR_0007s08150g [Populus trichocarpa] |
| 9 |
Hb_000307_160 |
0.0710605023 |
- |
- |
PREDICTED: uncharacterized protein At1g76070 [Jatropha curcas] |
| 10 |
Hb_001956_170 |
0.0724416621 |
- |
- |
hypothetical protein POPTR_0001s24950g [Populus trichocarpa] |
| 11 |
Hb_000028_090 |
0.0748223873 |
- |
- |
PREDICTED: uncharacterized protein LOC105135810 [Populus euphratica] |
| 12 |
Hb_009470_010 |
0.0778284349 |
- |
- |
- |
| 13 |
Hb_001401_130 |
0.0788939014 |
- |
- |
Calcium-binding EF-hand family protein, putative [Theobroma cacao] |
| 14 |
Hb_001386_070 |
0.0840204352 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1-like [Jatropha curcas] |
| 15 |
Hb_012180_070 |
0.090627549 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002186_190 |
0.0919435396 |
- |
- |
- |
| 17 |
Hb_000107_240 |
0.0945808766 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 18 |
Hb_009896_010 |
0.098040784 |
- |
- |
ABC transporter G family member 15 [Morus notabilis] |
| 19 |
Hb_000868_020 |
0.1020538885 |
- |
- |
- |
| 20 |
Hb_015183_090 |
0.1031612686 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 21-like [Jatropha curcas] |